Chromosomal-level genome provides an encyclopedia of animal evolution and adaptation to freshwater
Published in Ecology & Evolution

I once saw a video of sponges coughing—literally convulsing—to rid themselves of tiny ink particles. I was mystified and hooked. How could they do that without a nervous system or muscles? Sponges are one of the earliest branching animals, so understanding the underpinnings of their stimulus response could tell some fascinating stories about the evolution of nervous systems. The question of sponge sense has puzzled many researchers from diverse backgrounds, but making headway without a proper sponge genome for a readily accessible species was, to say the least, difficult.

With seed money from a joint program between Ludwig Maximilian University of Munich (Germany), and the University of Alberta (Canada), we began a sequencing effort.
This wasn’t easy. First, sponges are full of symbionts (mostly algae and microbes, but even worms, snails, and mites), so the isolation of only sponge DNA is difficult. Second, building a scaffold for genome assembly requires long intact DNA pieces, but something about sponge DNA makes it fall into little bits. During our first attempts, we burned through the sponge material I had collected and came out with a very poor sequence.

(photo: S. Leys)
Then there were two breakthroughs.
First, I met Joel Ussery, the manager of Resource Planning with Integrated Water Services in Victoria, British Columbia. One of the roles of his group is to coordinate research in the Greater Victoria Water Supply Area (20,550 ha of forested land owned by local government that surround the drinking water supply reservoirs). Learning what I was looking for, he asked if I’d like some sponges from the Head Tank, an enclosed reservoir just downstream of Sooke Lake Reservoir. Sponges? In the drinking water supply system?
The Head Tank is a large concrete chamber which holds the water before it goes into the eight kilometer underground concrete tunnel that sends the water to the distribution system in Greater Victoria. Every few years the Infrastructure Operations group headed up by engineer Matthew McCrank coordinates an inspection of the tank and tunnel. I was able to access the Head Tank and beginning of the tunnel during an inspection, and discovered their walls were coated in sponges!
One crucial aspect made them perfect samples for genome assembly: they lived in the dark and had no algal symbionts. Thanks to Joel, Matt, Jason Dales (the supervisor who coordinated the inspection), and their safety crew, I collected masses of them.

The second breakthrough was a timely meeting with Dr. Mansi Srivastava from Harvard University. Mansi has extensive experience with genome sequencing projects, and introduced me to Dovetail Genomics who offer high quality sequencing/assembly outputs. After a year (and not without hiccups), we had the sequence data.
As an international team of highly experienced and motivated collaborators, postdocs, and graduate students we discovered that, surprisingly, the genome assembly had captured nearly complete chromosomes.
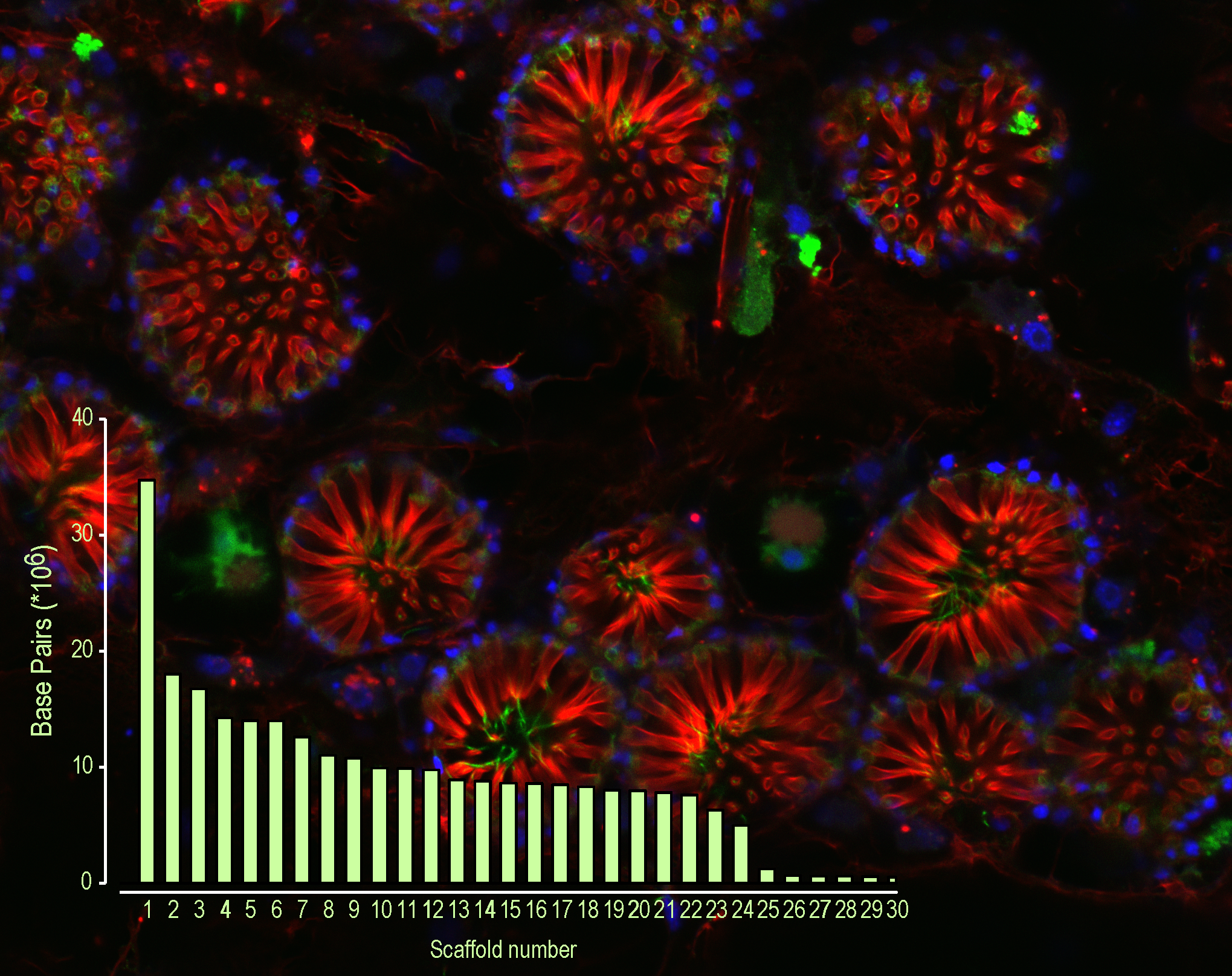
Such a ‘shiny’ genome leaves a lot to be explored. For a start, sponges have twice as many genes as most animals. Do some genes coordinate those coughs I saw earlier? Will others explain how the thin veneer of the sponge skin prevents loss of salts? Will some hold clues to surviving freezing temperatures?
We have already been able to use this research to show how much sponges resemble other animals genomically, even if they look very different morphologically. They have much the same regulatory systems, their genome is marked by similar methylation “tags”, and their genes have similar arrangements on the chromosome as many more recently evolved animal species.
There is much still to learn from this resource, and linking the genome to sponge ecology will be a good next step to understanding the “how” and “why” of sponge sense response.
For access to the resource and other information on freshwater sponges, please visit https://spaces.facsci.ualberta.ca/ephybase/
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in