Concurrent DNA hypomethylation and epigenomic reprogramming driven by androgen receptor binding in bladder cancer oncogenesis
Published in Cancer and Genetics & Genomics
Explore the Research
Concurrent DNA hypomethylation and epigenomic reprogramming driven by androgen receptor binding in bladder cancer oncogenesis
Click on the article title to read more.
Continued with our previous study [1], our group continues to explore the deeper mechanisms behind the occurrence and development of diseases. The intratumoral heterogeneity (ITH) of bladder cancer leads to treatment resistance and immune evasion, affecting clinical prognosis. Currently, the molecular and cellular mechanisms that promote the ITH in bladder cancer remain unclear.
Previously, we utilized the EpiTrace technology, which allows for the tracing of cell age and evolution [2], to perform single-cell temporal analysis on bladder cancer tissues at different stages [1]. We identified a TM4SF1-positive cancer cell subgroup (TPCS), which represents a critical node in the epigenetic evolution of bladder cancer, driving tumor development.
Bladder cancer shows a gender bias in its incidence and progression, with a higher occurrence rate in males compared to females. However, female patients often have a later stage at diagnosis and a poorer prognosis. Our team employed CUT&Tag sequencing technology to accurately map the trajectory of the androgen receptor (AR) in bladder cancer cells. We discovered that AR could drive the heterogeneity of tumor cells by altering DNA methylation patterns and regulating gene expression.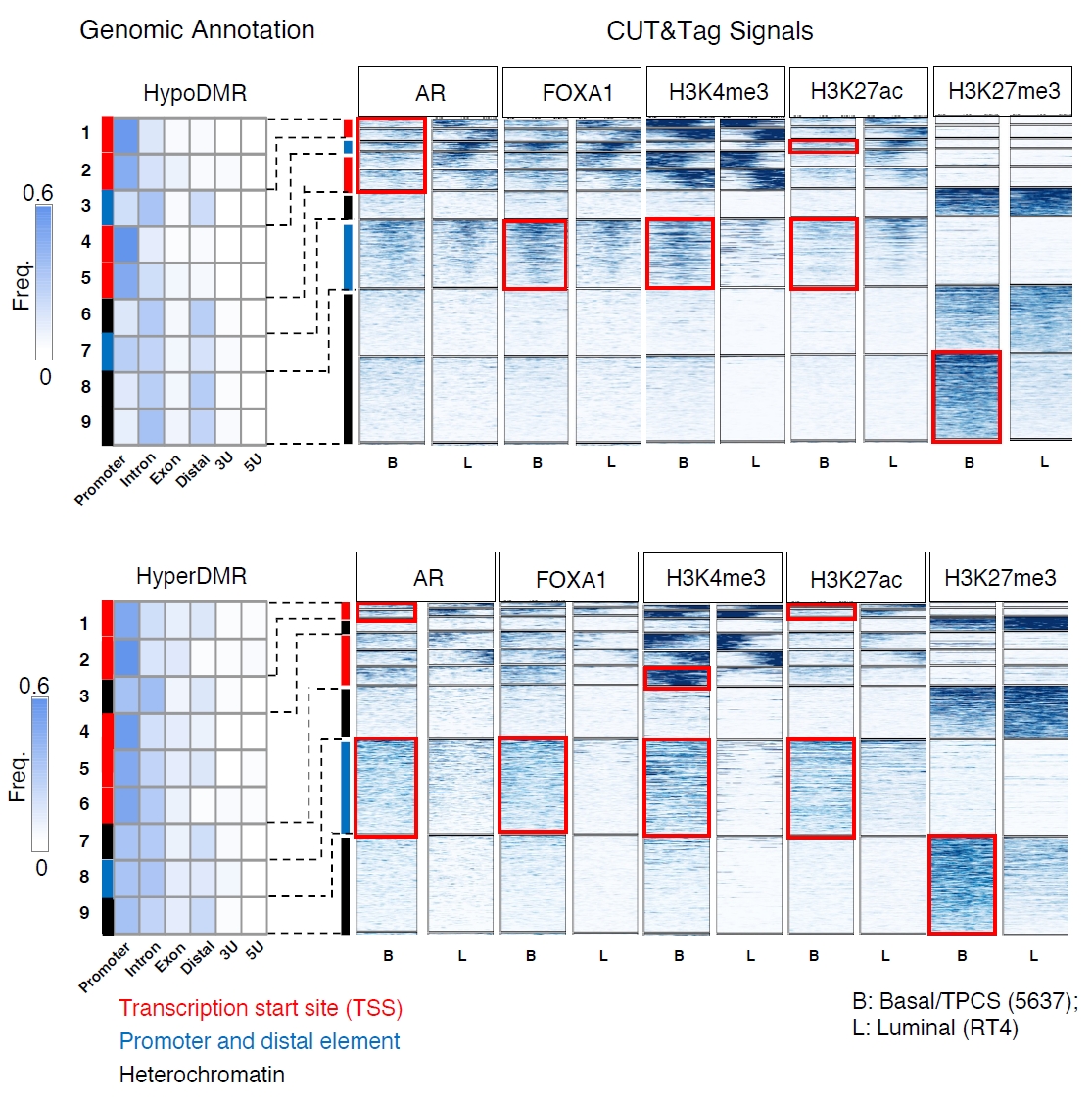
We selected two cell lines with different malignancy levels: RT4 (low malignancy) and 5637 (high malignancy), and through comparing these two cell lines, we further analyzed the interaction between the transcription factor FOXA1 and the androgen receptor AR. The study found that AR's gene regulation is not conducted independently but in close cooperation with other transcription factors (especially FOXA1), facilitating a complex mechanism of epigenetic reprogramming that promotes the evolution and heterogeneity of bladder cancer cells.
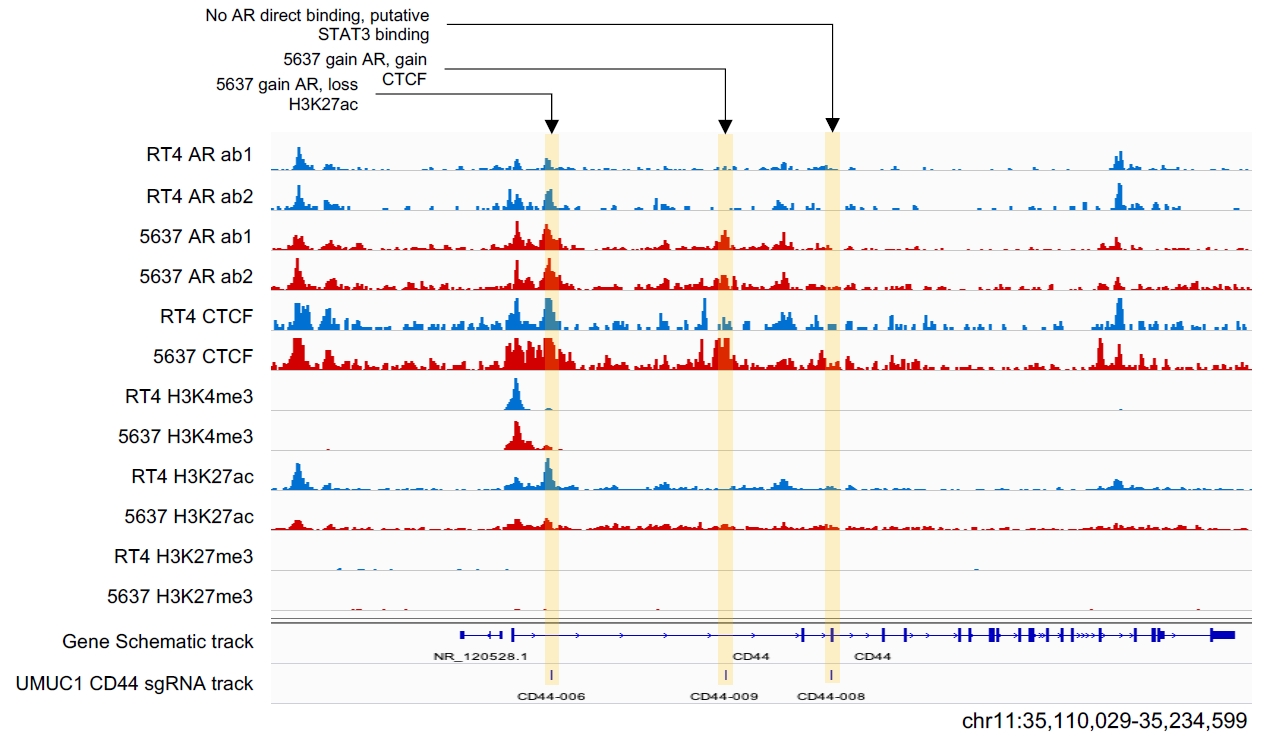
This research employed CUT&Tag sequencing technology to precisely map the "action trajectory" of the androgen receptor AR in bladder cancer cells for the first time, revealing the driving mechanisms behind the development of tumor heterogeneity and gender disparities.
References
[1] Xiao Y., et al. Integrative Single Cell Atlas Revealed Intratumoral Heterogeneity Generation from an Adaptive Epigenetic Cell State in Human Bladder Urothelial Carcinoma. Adv Sci, 2024, 11(24): e2308438.
[2] Xiao Y., et al. Tracking single-cell evolution using clock-like chromatin accessibility loci. Nat Biotechnol, doi: 10.1038/s41587-024-02241-z. Epub ahead of print (2024).
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in