Crowded dendrimers are worse inhibitors
Published in Bioengineering & Biotechnology
When it comes to designing multivalent biological inhibitors (scaffolds conjugated with multiple copies of an inhibitory ligand, or ‘decoy’), more is usually better: inhibitory complexes with a higher ligand valency usually have higher binding affinity, and make for better inhibitors. A study of a range of multivalent complexes designed to bind and block influenza hemagglutinin, reported last month in Nature Nanotechnology, now revealed that ligand spacing is also a crucial factor in determining multivalent ligand affinity. These findings highlight the need to fine-tune the size, shape and valency of multivalent complexes to optimize affinity and inhibition efficiency, which is of particular relevance when designing antiviral drugs.
The human-adapted influenza virus infects target cells by first binding to carbohydrate residues on the cell membrane via hemagglutinin, a viral surface protein. Strategies aimed at interfering with this interaction, for example by targeting the sialic acid binding site of hemagglutinin, have been developed in the past, but the inhibitory efficacy of these drugs is limited by their weak interactions, toxicity and immunogenicity.
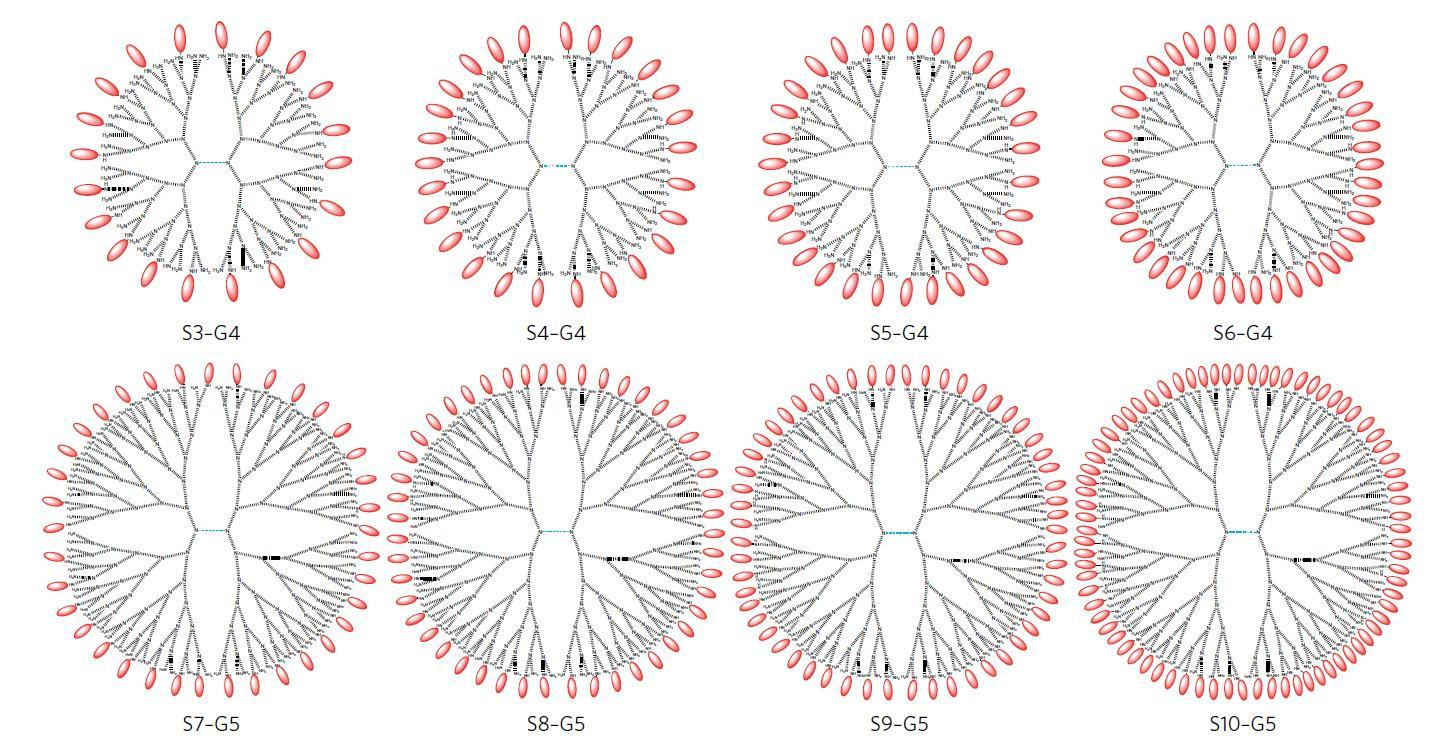
To address these issues, the authors of the study (Robert Linhardt and colleagues, from the Rensselaer Polytechnic Institute in Troy, New York), studied a range of different polyamidoamine (PAMAM) dendrimer scaffolds. PAMAM dendrimers are highly branched macromolecules of uniform and controllable size. By slow and controlled reductive amination, the authors synthesized several PAMAM dendrimer conformations conjugated at different stoichiometries with 6′-sialyllactose (6SL) — a soluble, non-toxic sialic acid that binds influenza hemagglutinin (Figure 1).
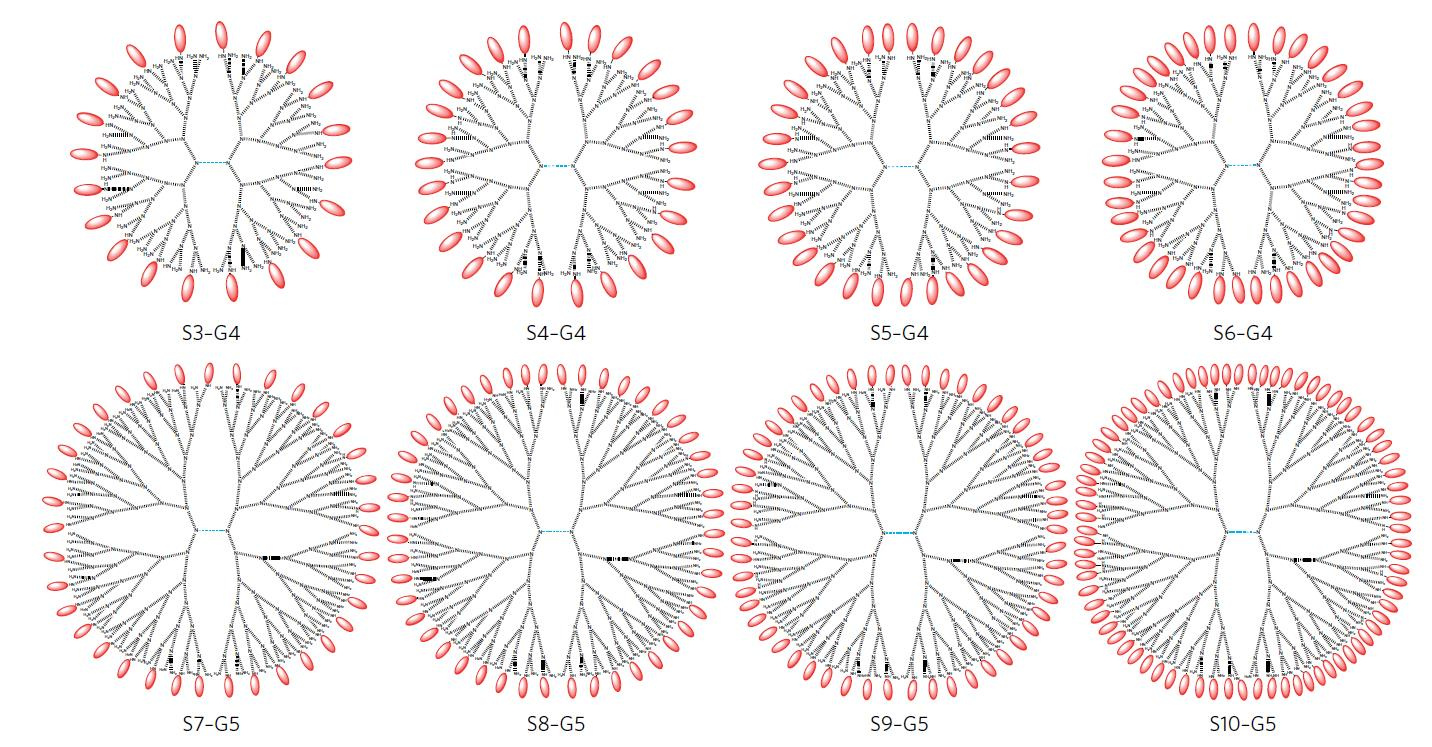
Figure 1. Range of S6L-dendrimer formulations synthesized. Reproduced from Figure 1, panel b, Kwon, S.-J. et al. Nat. Nano. doi:10.1038/nnano.2016.181, Springer Nature.
The authors’ analysis of the structure of hemagglutinin and of the different dendrimer complexes led to the identification of a critical factor determining inhibitor efficacy: conformations with an average distance between ligands of ~3 nm were the best inhibitors in infection assays in vitro, even when compared with configurations with higher valency (Figure 2). A dendrimer bearing twenty S6L ligands (named S3-G4, with an average distance between ligands of 3.1 nm) showed the highest affinity to hemagglutinin inhibitor in the panel, which also included complexes bearing up to 77 ligands.
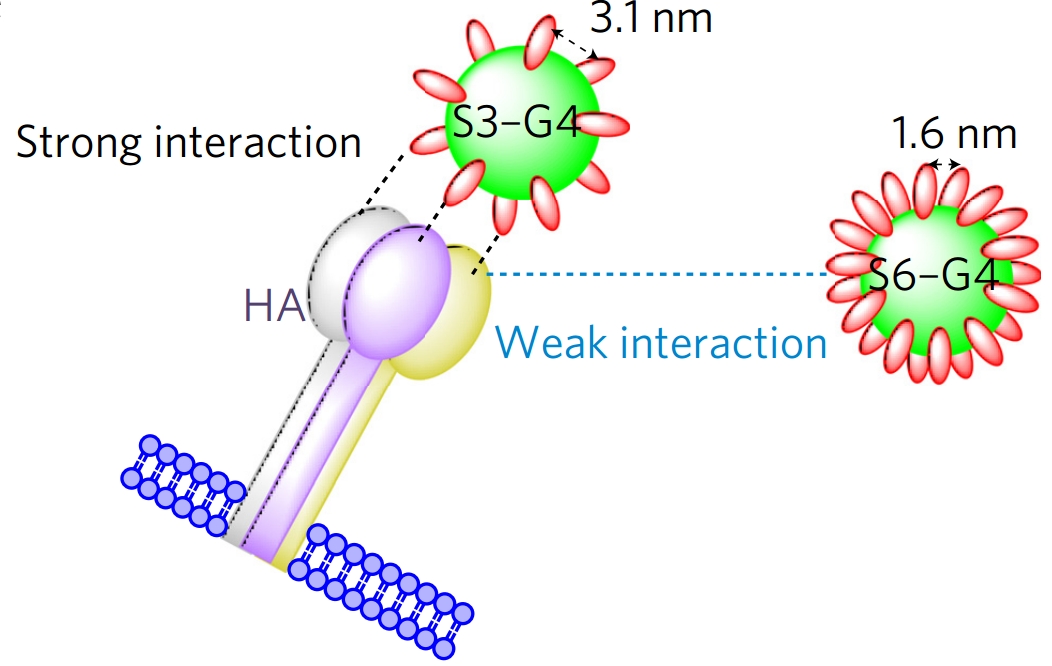
Figure 2. Effect of ligand distance in S6L-dendrimers. Reproduced from Figure 4, panel f, Kwon, S.-J. et al. Nat. Nanotechnol. doi:10.1038/nnano.2016.181 (2016). Springer Nature.
A set of in vivo experiments in which mice were infected with H1N1 confirmed the inhibitory efficacy of the S3-G4 complex. The majority of mice treated with the complex survived an usually lethal viral dose, and had lower viral titers in the lung, fewer tissue lesions and lower levels of inflammatory cytokines when compared with control mice infected with the same H1N1 strain. The results point to S3-G4 having a protective effect against influenza infection in mice through its high affinity for hemagglutinin.
The reasons why S3-G4 performed well were attributed to the structural properties of the new inhibitor, namely that ligand distance was optimized for hemagglutinin binding, which led to superior affinities than those of tightly arranged, higher valency complexes. Because of its higher affinity to the viral protein, the complex had potent therapeutic properties in vivo, highlighting the importance of optimal placing of inhibitory ligands on the dendrimer.
Highlighted research paper:
Kwon, S.-J. et al. Nanostructured glycan architecture is important in the inhibition of influenza A virus infection. Nat. Nanotechnol. doi:10.1038/nnano.2016.181 (2016).

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in