Environmental chlamydiae, the hot topic at an imaginary party
Published in Microbiology

Pangenomics reveals alternative environmental lifestyles among chlamydiae
https://www.nature.com/articles/s41467-021-24294-3
Let’s be honest, the first thing that pops into the head of most people when hearing “Chlamydiae” is the sexually transmitted disease caused by one species, Chlamydia trachomatis. However, in Matthias Horn’s group in Vienna, Austria, we are interested in the “other” chlamydiae. So I’ve had the same conversation many times, be it with family members, friends, hairdressers, or people you meet at a social gathering (pre-COVID).
Let’s say I am at an imaginary party (kinda what we have been doing the past 1.5 years) and have a conversation with my imaginary friend called Sam. I tell Sam that I am studying chlamydiae and Sam’s instant reply is of course: “The tripper?”. I respond: “No, you know, chlamydiae infecting humans are just a tiny fraction of this actually large group of bacteria. I don’t ignore them, but I focus on the other chlamydiae, which are likely thousand times more diverse.”
“Wow”, Sam responds, “so you are saying that the chlamydiae you are studying are actually the vast majority?”. Yes, that’s what I’m talking about. Sam: “But why are they important?”. Damn you, Sam.
So I go for the long explanation: “Our chlamydiae of interest are often collectively referred to as environmental chlamydiae, as many of the cultured representatives have been isolated from environmental samples like freshwater or soil. Like their human pathogenic relatives, these chlamydiae, however, depend on a eukaryotic host cell for replication, i.e., they are obligate intracellular symbionts. So, by studying environmental chlamydiae, we can learn about the evolution of microbe-host relationships and the emergence of pathogens. For most cultured environmental chlamydiae, the hosts of choice are unicellular eukaryotes - protists - that can be found almost everywhere in the environment. These protists are so abundant that they likely make up twice as much biomass on earth as all animals taken together.”
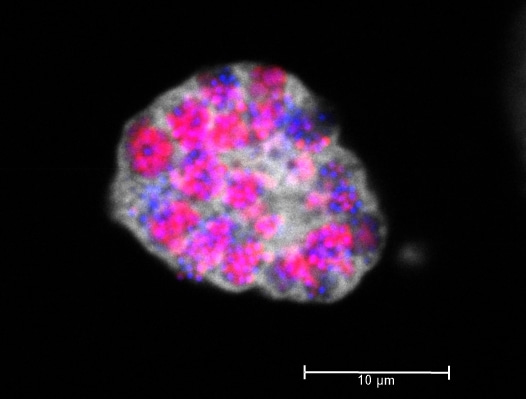
"Look at that these cute Parachlamydia acanthamoebae UV7 cells in their protist host Acanthamoeba castellanii, Sam. My good friend Angelika Schwarzhans took this pic."
But here’s the thing, honestly, we know very little about most chlamydiae, except that they are present in many environments based on marker gene (16Sr RNA) surveys and we think they might more often than not be associated with protists. Things I don’t have to worry Sam about.
Sam seems somewhat interested and asks: “OK that’s pretty cool, what have you found out about those chlamydiae?”. I tell Sam about my great collaborators Frederik Schulz and Sean Jungbluth from the Joint Genome Institute who identified 82 chlamydial metagenome-assembled genomes (MAGs) in the Genomes from Earth’s Microbiomes (GEM) catalog. The environments they come from range from the world’s oceans to forest soils. I tell Sam that we almost couldn't believe our eyes when we calculated the first phylogenomic tree including those MAGs. Not only are there so many new genomes, but they are also incredibly diverse. “We actually almost doubled chlamydial diversity, adding many new species and genera, and even seven new families. We called them MCF-A to MCF-G”, I explain, almost spilling my imaginary beverage out of enthusiasm.

"Look at that added chlamydiae diversity, Sam."
Sadly, Sam is not impressed by phylogenetic diversity and is not very interested in taxonomic units. But that’s ok, I’m used to it. “What might be interesting”, I try to save the conversation, “is that some of the new genomes we found were quite distant from any chlamydiae known so far. Others are closely related to well-known chlamydiae, but still seem to be quite different. Anyways, what they all have in common is that they possess the genomic features necessary to live as intracellular symbionts like all known chlamydiae. That is reassuring, so likely they are all very intimate with a symbiotic partner”. Sam seems quite amused and giggles: “Partner, more like a victim”. Fair enough Sam, you are an odd one.
“But listen to this”, I feel like I really got Sam hooked now, “in one of the best known clades, the Parachlamydiaceae, we were in for quite the surprise when my epic colleague Astrid Collingro found genes for harvesting light to produce energy in their genomes. So chew on that Sam, some chlamydiae went “green”, figuratively speaking, and can likely use light as an energy source. Might be to survive in the antarctic lakes they live in or maybe this is a beneficial feature for their host? We don’t really know yet, but isn’t that exciting?”
There was no stopping me: “Apart from this anomaly we were interested in a recently described feature of some chlamydiae, the ability to live in anoxic environments. Amazing Tamara Halter and I went for a data dive to identify genes involved in anaerobic energy production. And to our surprise we found that pyruvate fermentation to acetate is widespread in many chlamydiae, followed by the arginine deiminase pathway. And there might be additional lineages capable of producing hydrogen.”
Sam seems a bit overwhelmed, but correctly summarizes: “So in essence you and your colleagues found that chlamydiae are generally more flexible when it comes to oxygen than what was previously thought”. “Exactly”, I happily agree and add “In a way, this actually makes a lot of sense, as one of the best-known hosts of environmental chlamydiae, Acanthamoeba castellanii, is capable of thriving in anoxic environments. So they can be buddies even in sediments under low (no) oxygen conditions.” Sam giggles again: “Takes my breath away” and coughs, hiding his lame pun. Sam, you are an odd one indeed, but that’s why we are imaginary friends.
“If you want to know more about those chlamydiae and the environments in which you can find them, I can send you the link to the paper”, I try to convince Sam, but get interrupted, “Thanks Stephan, I’m good, enough chlamydia for one night”.
By Stephan Köstlbacher
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in