Gene gain in intracellular bacteria or how Chlamydiae defy the odds
Published in Microbiology

People say that solitude makes you stronger and to a degree this seems intuitive. Alone you have to tackle every issue that comes your way by yourself and need to have or acquire the tools leading to success. In reality, human character is not so simple, but this principle generally applies to free-living bacteria. Bacteria that live freely in the environment need to produce all the necessary building blocks missing from their environments to survive, grow, and divide. But these bacteria can also obtain DNA from other microorganisms in the large and complex communities they tend to live in. These horizontal gene transfer (HGT) events are the main way bacteria acquire new traits (Figure 1). Together, this can lead to relatively larger genomes (with exceptions), which encode complete metabolic pathways, defense systems etc.
But not all bacteria live alone. Many live in long-term and tight associations with other organisms as symbionts. Some even live within the cells of another organism, so-called endosymbionts. Many endosymbionts are hosted by eukaryotes, such as us humans which basically are a humongous pile of eukaryotic cells. Inevitably this makes us attractive hosts, but the bad news: not all symbionts are beneficial. Prominent examples of bacterial endosymbionts that are parasites or pathogens from history and present day life are those that cause sexually transmitted infections like gonorrhea, syphilis, or a personal favorite, chlamydia.
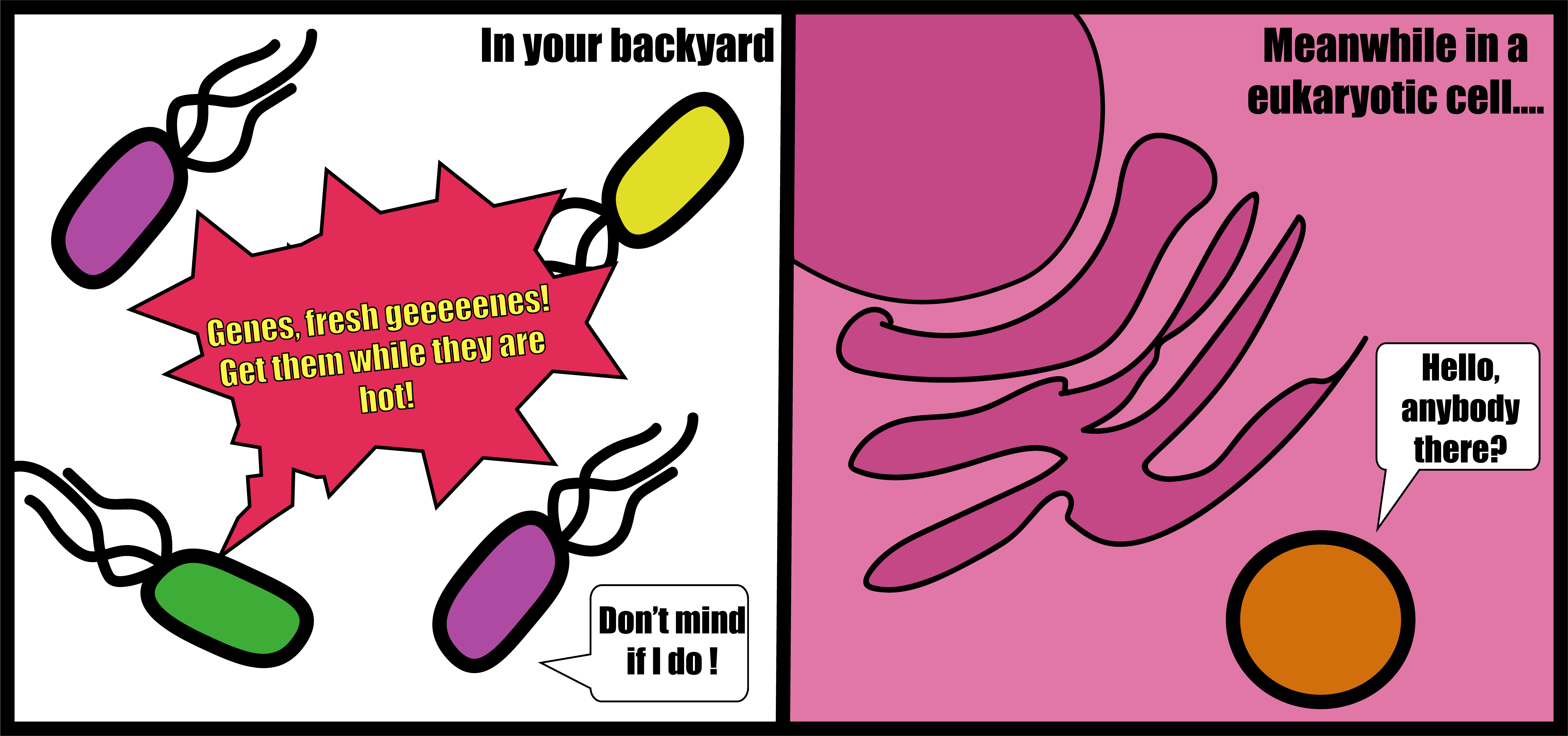
Figure 1: Opportunities for gene exchange differ for free-living and endosymbiotic bacteria.
How does the life of an endosymbiont compare to a free-living bacterium? Let's assume the case of a bacterium that lives happily in a host cell that does not mind the (uninvited) guest. Needless to say, the following example will be a drastic oversimplification. With an abundance of nutrients in the host cell, the bacterium does not need to produce many of its own building blocks. With lowered selection pressure to keep genes for producing nutrients they will tend to be lost. Over time the bacterium will become incapable of living outside without the nutrient supply from the host. While life is comfortable, the bacterium gets lazier and co-dependent, which is fine as long as it always has a host around. But a second issue aggravates the situation. Confined to a small space, only a small number of endosymbionts can live in each host cell and only few offspring will end up transmitted to new host cells. Of course the bacterium is evolving and acquires new mutations. It might break your heart to read this, but new mutations tend to be neutral or slightly negative rather than positive. This is bad news for the bacterium because when only a small number of offspring move on to a new host cell, it leads to negative mutations piling up and can result in “unhealthy” genomes. Negative mutations can in principle be fixed by acquiring a “healthy” version of a gene by HGT from another bacterium and in larger populations “survival of the fittest” means that those with an unhealthy gene will simply be outcompeted. However, endosymbionts have small population sizes and are shielded from the outside world. Thus, the bacterium has few opportunities to exchange genes with other bacteria (Figure 1), leading it down the road to a small and “unhealthy” genome. Poor bacterium, genome decay is sad. But it seems that some fascinating endosymbionts have defied genomic decay.
As mentioned before, Chlamydia species are well known pathogens and endosymbionts. They belong to the larger phylum Chlamydiae which, as far as we know, consists exclusively of strict endosymbionts. A plan was made: to reconstruct the billion year long evolutionary history of this phylum and find out what happened with respect to genome evolution. The teams of Thijs Ettema at then Uppsala University (Sweden), now Wageningen University & Research (the Netherlands), and Matthias Horn at the University of Vienna (Austria) decided to tackle the challenge together. While chlamydiae are best known for human pathogens, most are actually hosted by unicellular eukaryotes - protists - found everywhere in the environment. Protists are very abundant and make up more than twice the biomass of all animals together. Many protists are predators that eat microbes. Protist endosymbionts can obtain DNA from the bacterial prey the host eats or other endosymbionts they share a host cell with (which is not uncommon). Together with larger population sizes and easier transmission in protists this could explain why chlamydiae that infect protists have up to three times larger genomes than human pathogen chlamydiae. We were very curious to study how chlamydiae genomes evolved in light of the complications of life as a symbiont and discrepancies in genome sizes.
Jennah Dharamshi and Stephan Köstlbacher joined forces to collect a large and diverse set of genomes to reconstruct Chlamydiae evolution. By modeling the evolution of all chlamydial genes with the help of Max Emil Schön and Astrid Collingro, we could reconstruct the genes in the last common ancestor of all chlamydiae (LCCA). We confirmed that LCCA did indeed already possess the traits for endosymbiotic life. But more interesting was another observation. Previously, it was speculated that LCCA had a relatively big genome, like the protist-infecting chlamydiae we know today, and that human pathogenic chlamydiae had small genomes due to gene loss. But we observed something completely different. The genome size, measured as the reconstructed gene number, in LCCA was actually not much larger than that of today's human pathogenic chlamydiae with their small genomes.
This was quite the puzzle. We started to one by one reconstruct the gene content of chlamydiae from the LCCA to the protist-infecting chlamydiae today that have the largest chlamydial genomes. And we came across a surprising finding: Ancestors with small genomes were getting bigger over time, almost doubling their gene content. To our knowledge, and in light of a tendency towards genome decay in endosymbionts, this was unheard-of (Figure 2). When tracing back where genes gained in these ancient chlamydiae came from, we observed HGT from many different bacteria. Interestingly, many gained genes could be tied to functions related to adaptation to high levels of oxygen and energy generation. While the ancestor of all chlamydiae was adapted to low oxygen conditions, protist-infecting chlamydiae became better at dealing with high oxygen, which can be toxic if you don’t have the right genes.
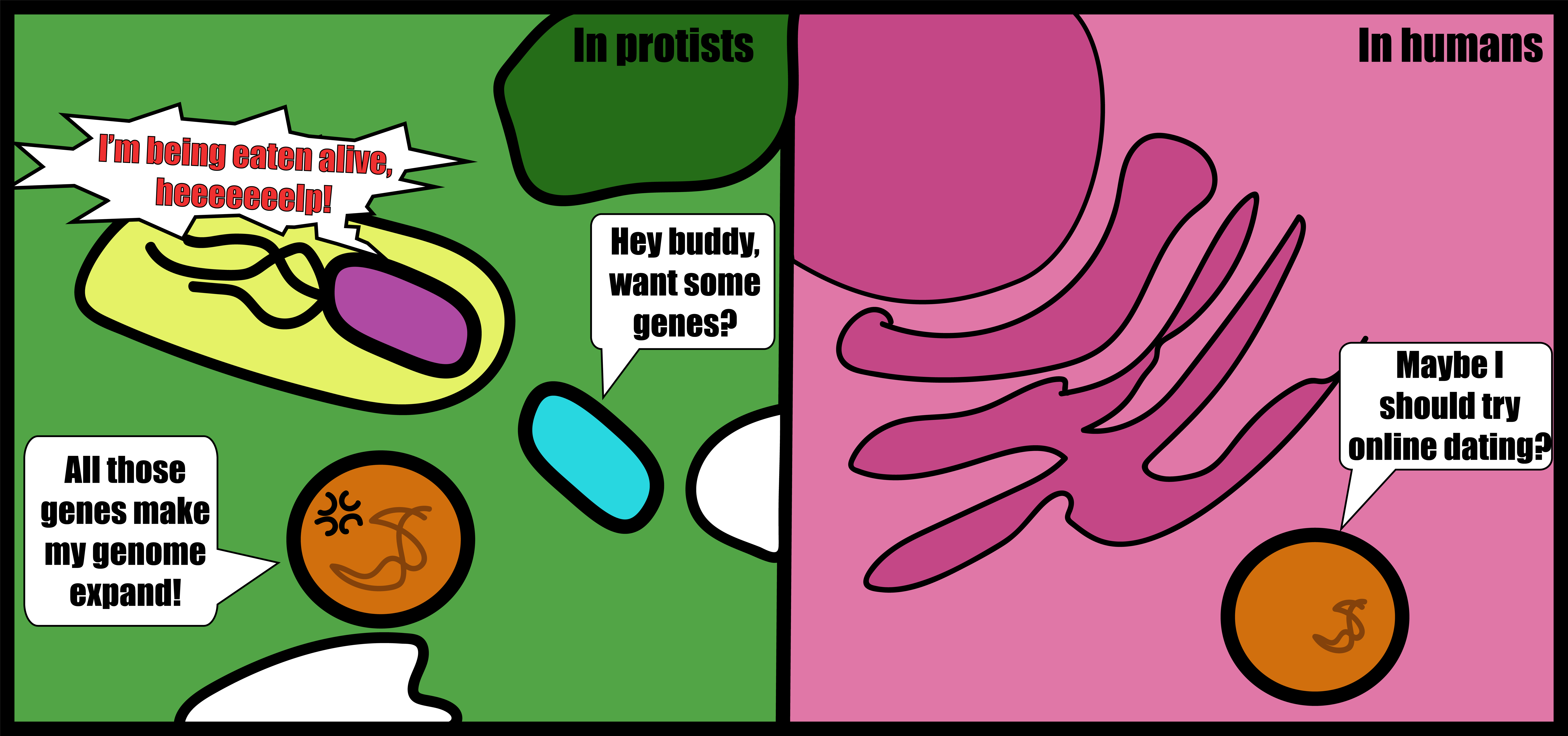
Figure 2: How the model of genome evolution in human pathogenic chlamydiae differed from what we observe in protist-infecting chlamydiae.
Protist-infecting chlamydiae seem to defy what we know about genome evolution in endosymbionts. Instead of maintaining or reducing their gene content over time, some protist-infecting chlamydiae drastically increased their gene content. Like many, we had also thought that solitude makes you stronger, but chlamydiae showed us that even while depending on others you can defy the odds. If you want to get even more inspired by Chlamydiae, have a look at our paper.
By Stephan Köstlbacher and Jennah Dharamshi
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in