How do DNA Replication Origins work?
Published in Physics, Cell & Molecular Biology, and Genetics & Genomics
In this paper, we inquire about the role of DNA sequences in determining their own replication behavior, without proteins' help, relevant to prebiotic scenario, where no such external help was available. This question is important, for, an answer would instruct us in rationally altering the origin sequence to alter the origin functionality through genetic engineering tools, thus modifying the DNA replication dynamics, irrespective of the presence or absence of interpreting protein molecules.
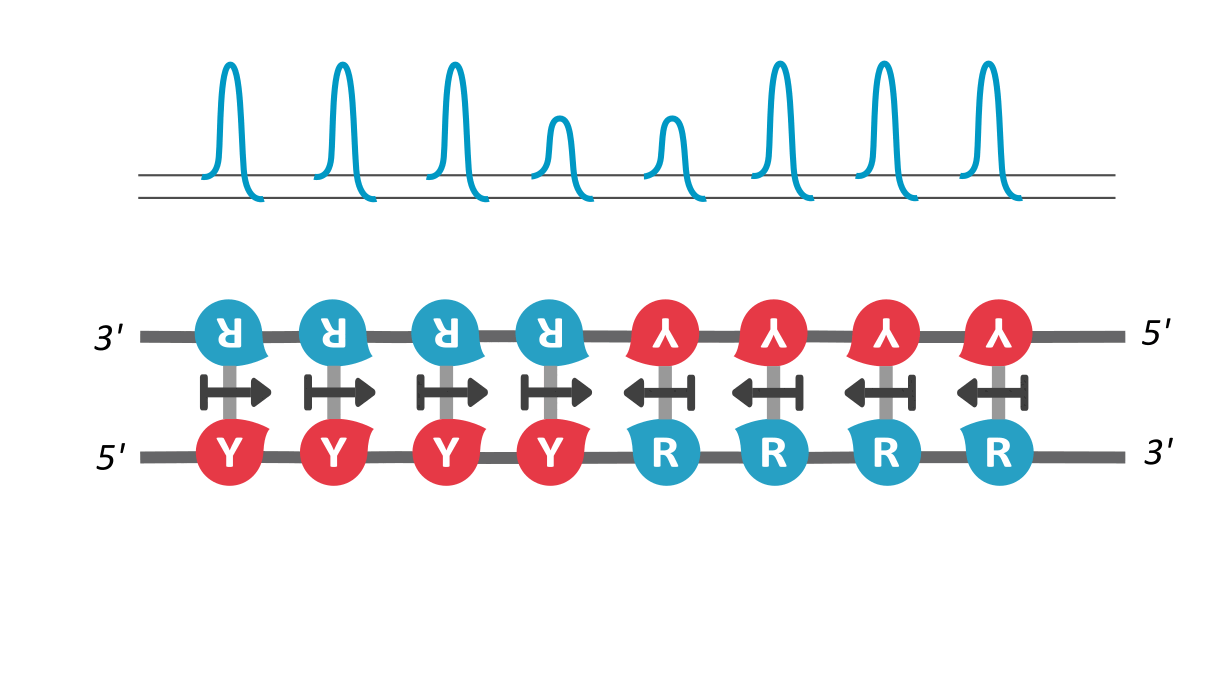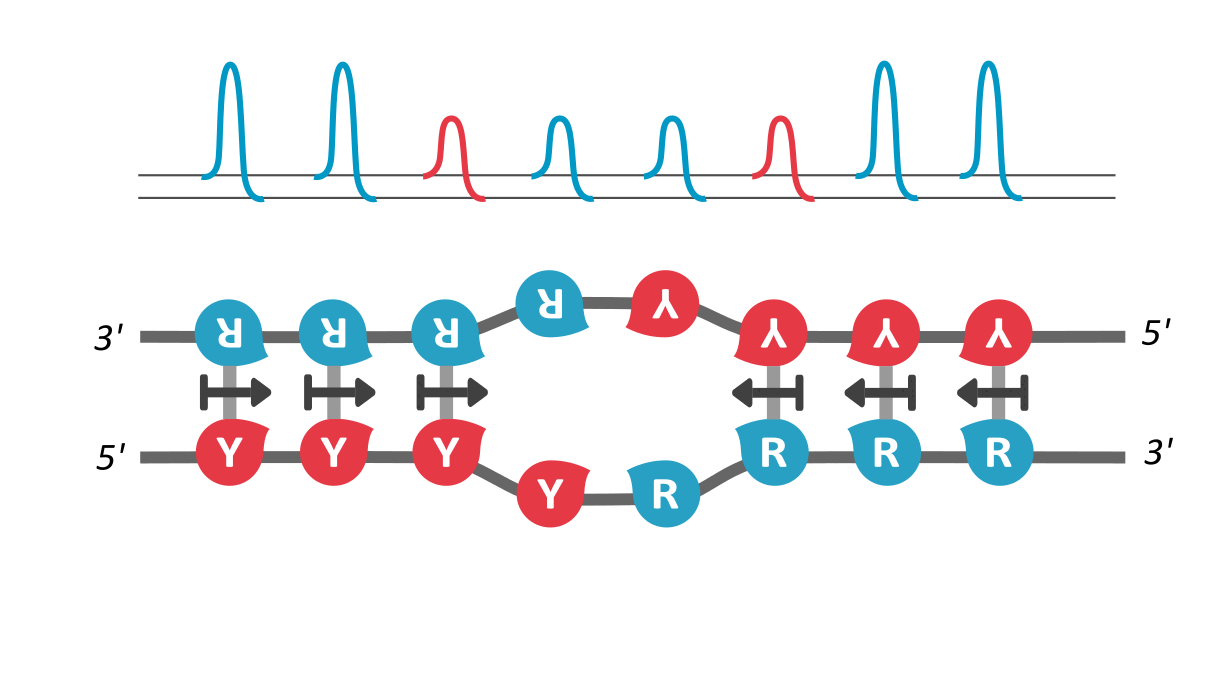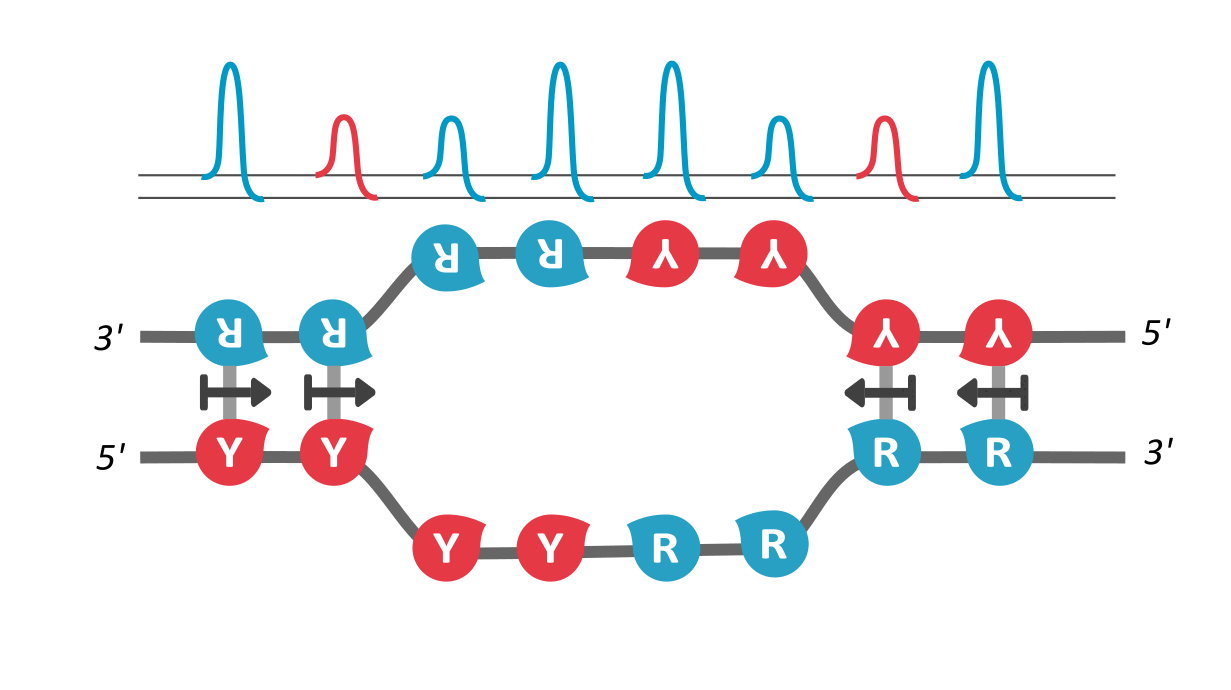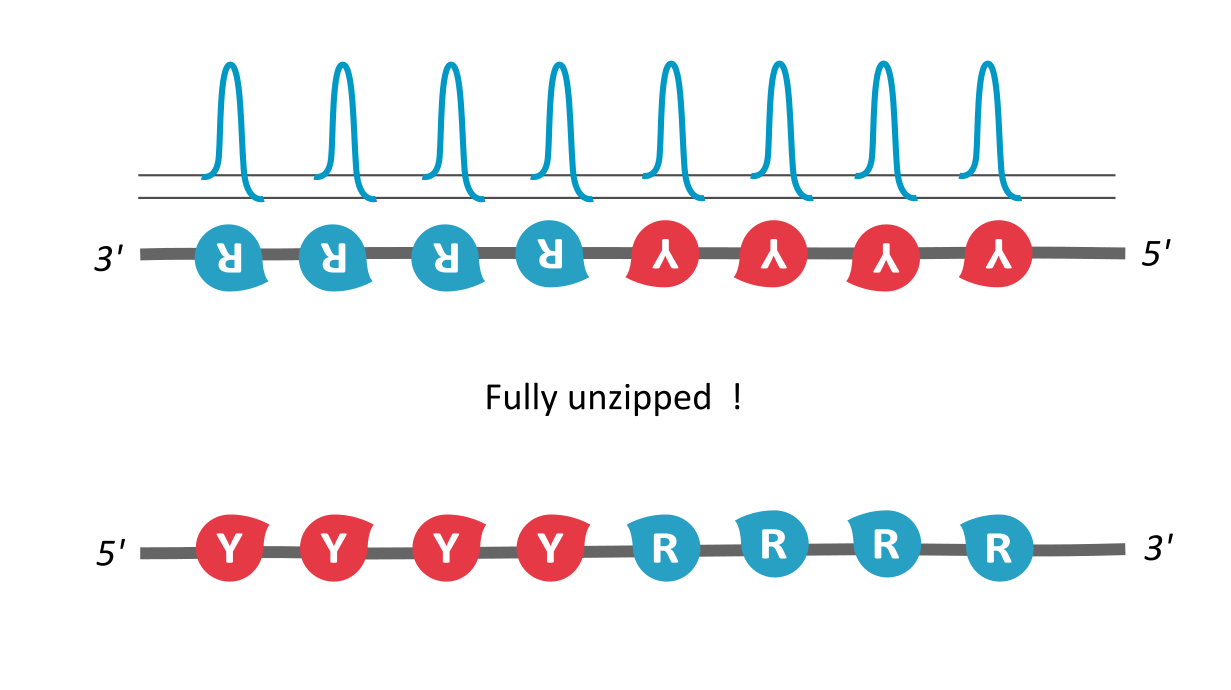
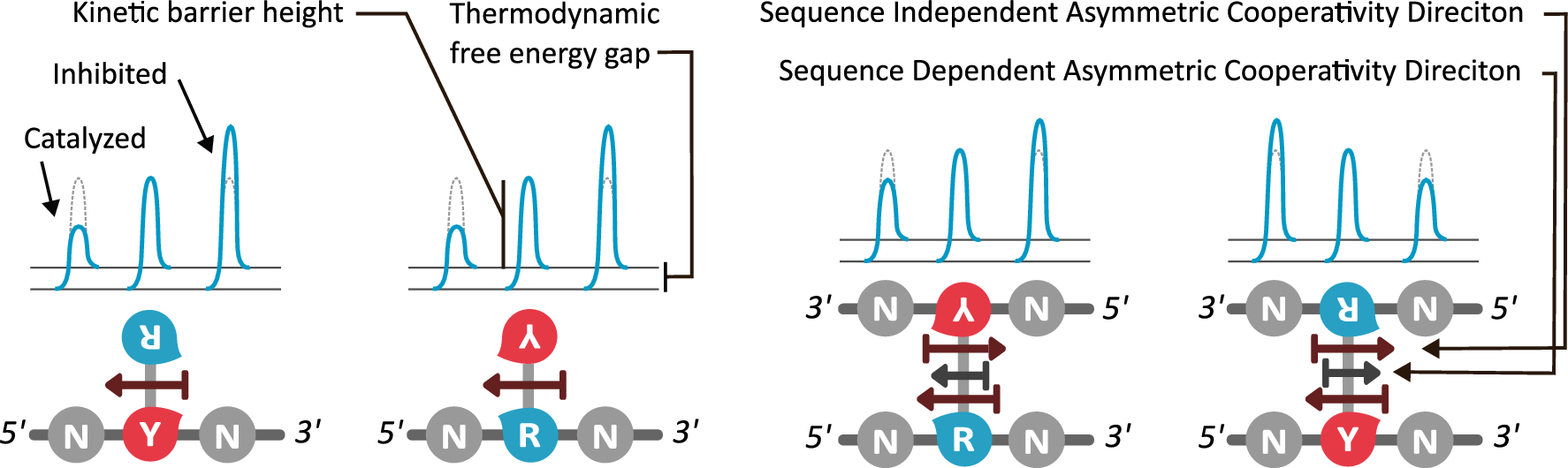
In our paper, we utilize the proposed existence of a kinetic property that helps tie the DNA sequence with its local unzipping rates, helping us convert time-independent symbols to time-dependent dynamics. This property, called asymmetric cooperativity, has been proposed earlier to make sense of the counter-intuitive properties of DNA, like its unidirectional replication and anti-parallel strand orientation, which were shown to be evolutionarily beneficial for the DNA/RNA (Link, link). Since the nature of sequences dictate their unzipping propensity, our goal simply is to evaluate the unzipping rates of all possible sequences of a certain length, in the presence of asymmetric cooperativity, and find the character of the sequence(s) that maximize the unzipping/replication rate.
We find that RY-palindromic sequences, where the 3'-end is loaded with purines (R) and 5'-end, with pyrimidines (Y), have the fastest unzipping rates, simply because they have low kinetic barrier at the purine-pyrimidine interface, (See attached figure). This facilitates rapid and efficient strand separation, and makes such sequences potential replication origins. Our study provides bioinformatics evidence supporting this claim, by identifying skewed palindromic regions at known replication origins across multiple biological domains, including mitochondrial DNA, bacteria, archaea, and plasmids. This preferential unzipping of origin sequences can also direct the replication machinery towards the unzipped regions, thus avoiding the time-consuming search for structural origin signatures along the sequence, by the replication machinery.
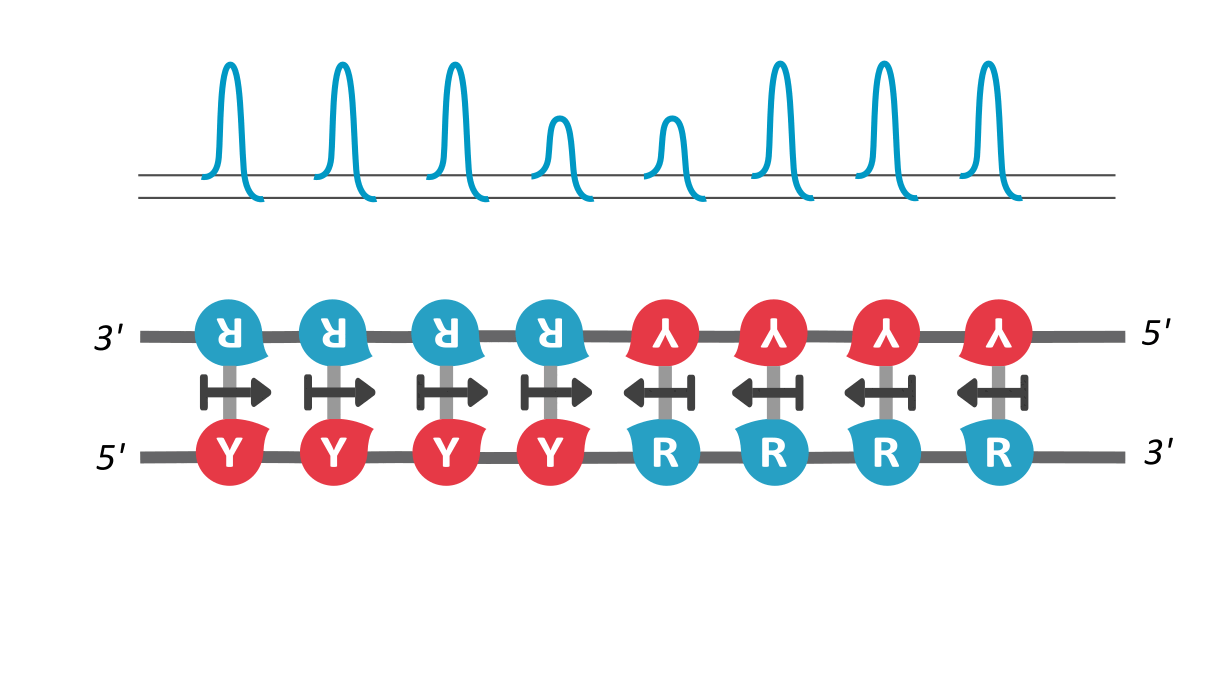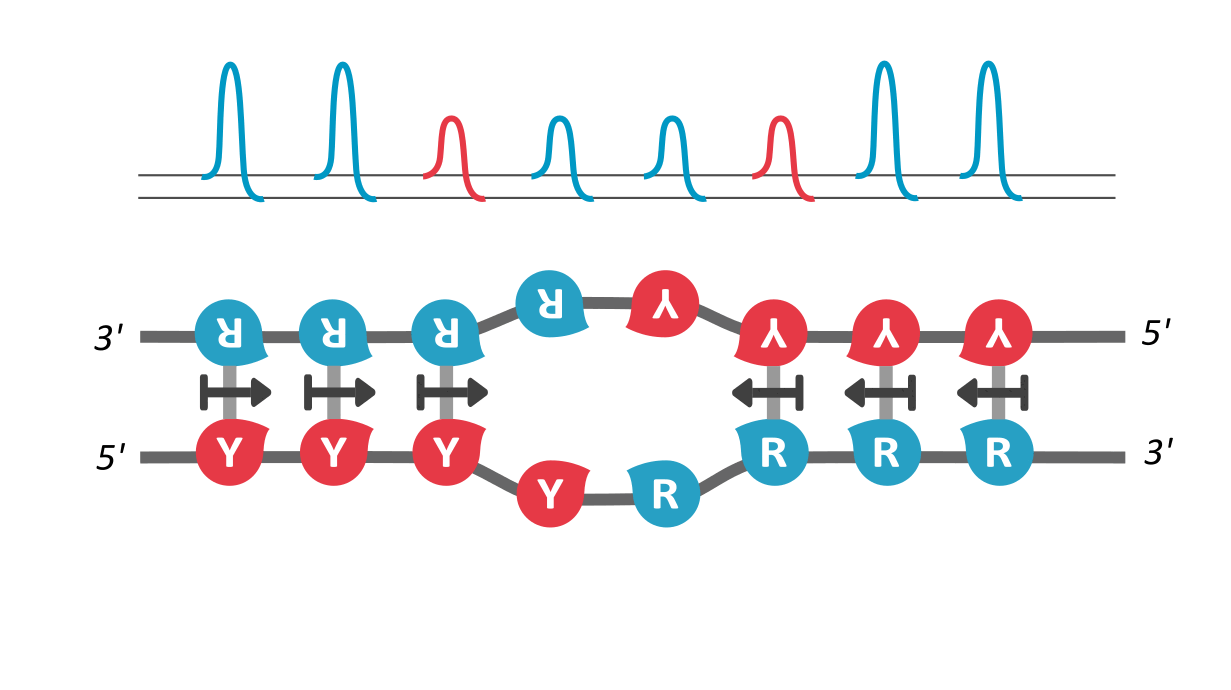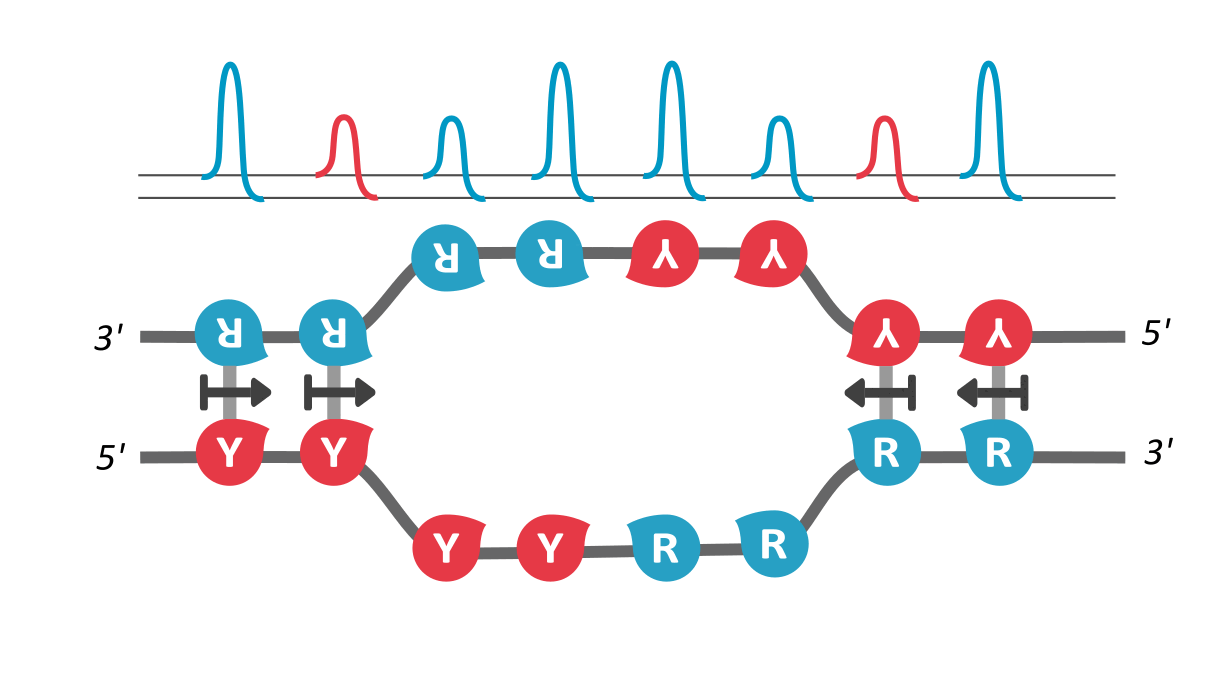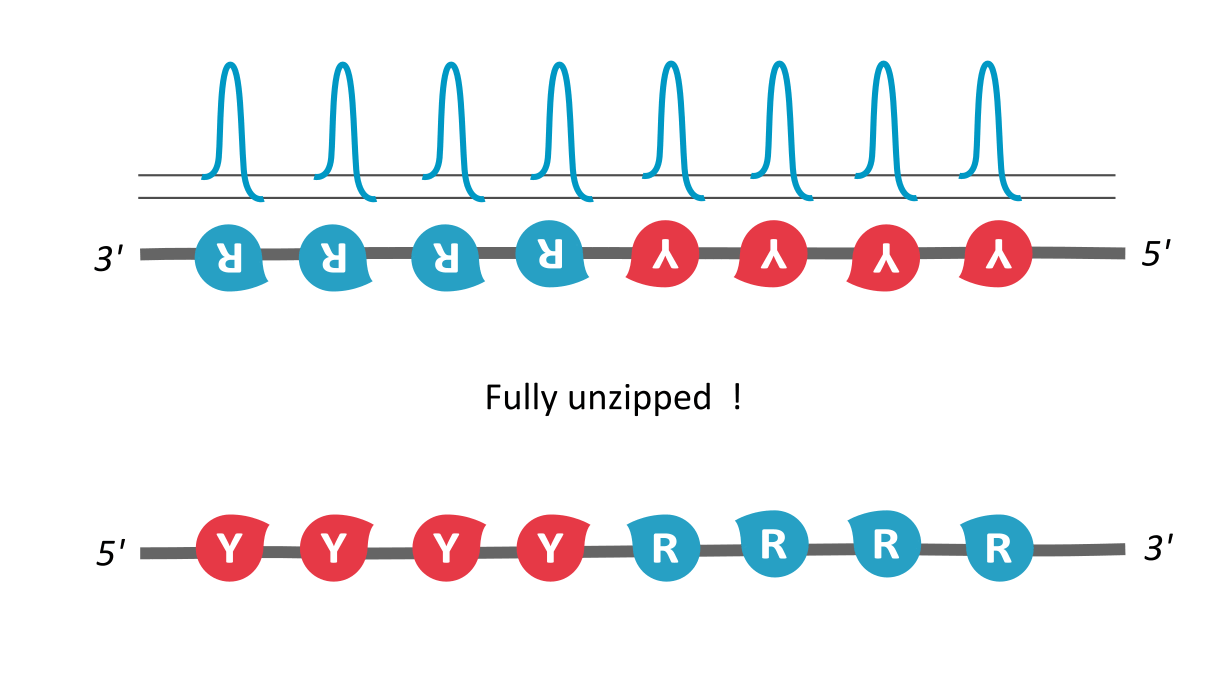
palindromic sequence 5′-YYYYRRRRR-3′ functioning as a replication origin
site
There is another interesting way to verify our results: According the asymmetric cooperativity model, if the two arms of the palindromic sequence, ones with purines and the other with pyrimidines, are swapped, the pyrimidine-purine interface would have high kinetic barrier, and thus will cease to function as an origin. We found in the literature precisely such an experiment, performed on the mitochondrial light strand origins, where, such swapping was found to completely abrogate the origin functionality (Link). This experiment precludes a common explanation for origin functionality of palindromes, that of their stem-loop formation potential, for, the swapped-arm sequences are also palindromes.
The main breakthrough here is connecting the sequence with its unzipping/zipping kinetics, using the kinetic property of asymmetric cooperativity. This connection became possible by pivoting to kinetic, as opposed to thermodynamic, interactions between neighboring base pairs in DNA. Such a connection not only leads to an explanation for the replication origin, but can be shown to dictate the overall replication dynamics as well, through the well-established observation of GC-skew in the genomes of both prokaryotes and eukaryotes . By demonstrating the ability of DNA to influence its own replication kinetics through modification of its sequence, we arrive at a much more interesting conclusion, that evolution can directly maximize the replication potential of sequences through natural selection, by modifying sequences, without any enzymatic intermediaries, thereby kickstarting prebiotic evolution.
Follow the Topic
-
Journal of Molecular Evolution

This journal covers experimental, computational, and theoretical work aimed at deciphering features of molecular evolution and the processes bearing on these features.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in