Influenza vaccines—not all about neutralization
Published in Bioengineering & Biotechnology
The identification of which pathogen molecules are recognized by the immune system following vaccination, particularly in terms of quantifying changes in the antibody repertoire, has been technically challenging, in no small part because of the wide range of immune specificities found in each individual. Progress has been made in this area by analyzing circulating B cells (the cell type that secrete antibodies) or through traditional serologic quantification of circulating antibodies. However, these methods fail to capture dynamic changes in antibody repertoires and lack the capacity for functional characterization of each antibody clone within the multitude of antibodies generated after vaccination. A new report by George Georgiou (University of Texas) and colleagues, published this November in Nature Medicine, describes how high-resolution proteomics analysis of antibodies can be combined with high-throughput sequencing of B cell receptor transcripts to pinpoint changes in the antibody repertoire of individuals vaccinated with a seasonal (IIV3) influenza vaccine.
Every year, healthcare programs throughout the world coordinate the vaccination of millions of people with ‘seasonal’ influenza vaccines – a mix of inactivated influenza virus strains selected for their prevalence in the general population. This type of multipronged approach primes the immune system against the predominant influenza variants, and is efficacious in the majority of the population. But this protection is far from being complete: seasonal flu vaccines have low efficacy in young adults and, more worryingly, in infants and the elderly, which are at greater risk of developing complications from influenza infections.
The reasons why influenza immunizations offer incomplete protection are not fully understood. One big factor behind this lack of efficacy is the antigenic variation that occurs in circulating virus strains, particularly at the level of hemagglutinin (HA) and neuraminidase (NA), two influenza surface proteins critical for the virus infection cycle. Variation in these and other viral proteins enable the virus to escape host defenses. To counter this viral escape mechanism, current seasonal influenza vaccines contain particles from several strains to try to cover as much of this variation as possible. The trivalent vaccine currently in use, for example, contains inactivated viral particles from influenza A H1N1 and H3N2 strains, and one influenza B strain.
In contrast to viral antigen variation, other factors influencing the efficacy of influenza vaccines might be associated with the host. Vaccines act differently among individuals, particularly in terms of the antibody response elicited. Ideally, the objective of vaccination is to generate antibodies that neutralize a broad range of viral protein variants (so-called broadly neutralizing antibodies), so that each individual can become immunized and respond to a wide range of viral strains.
In their new paper, Georgiou and colleagues determined the IgG clonotypes before and after vaccination by using a high-resolution liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS) in a proteomics pipeline to analyze antigen-specific antibodies in human serum (Figure 1). When comparing pre-existing and vaccine-elicited antibodies, it became clear that the majority of the response to vaccination is due to expansion of pre-existing antibodies. This observation highlights the significance of pre-exposure to the virus (that is, previous infections) in defining the response to vaccination; more specifically, previous exposures to influenza will decrease the diversification of the response to vaccination and may lead to weaker protection against divergent virus strains.
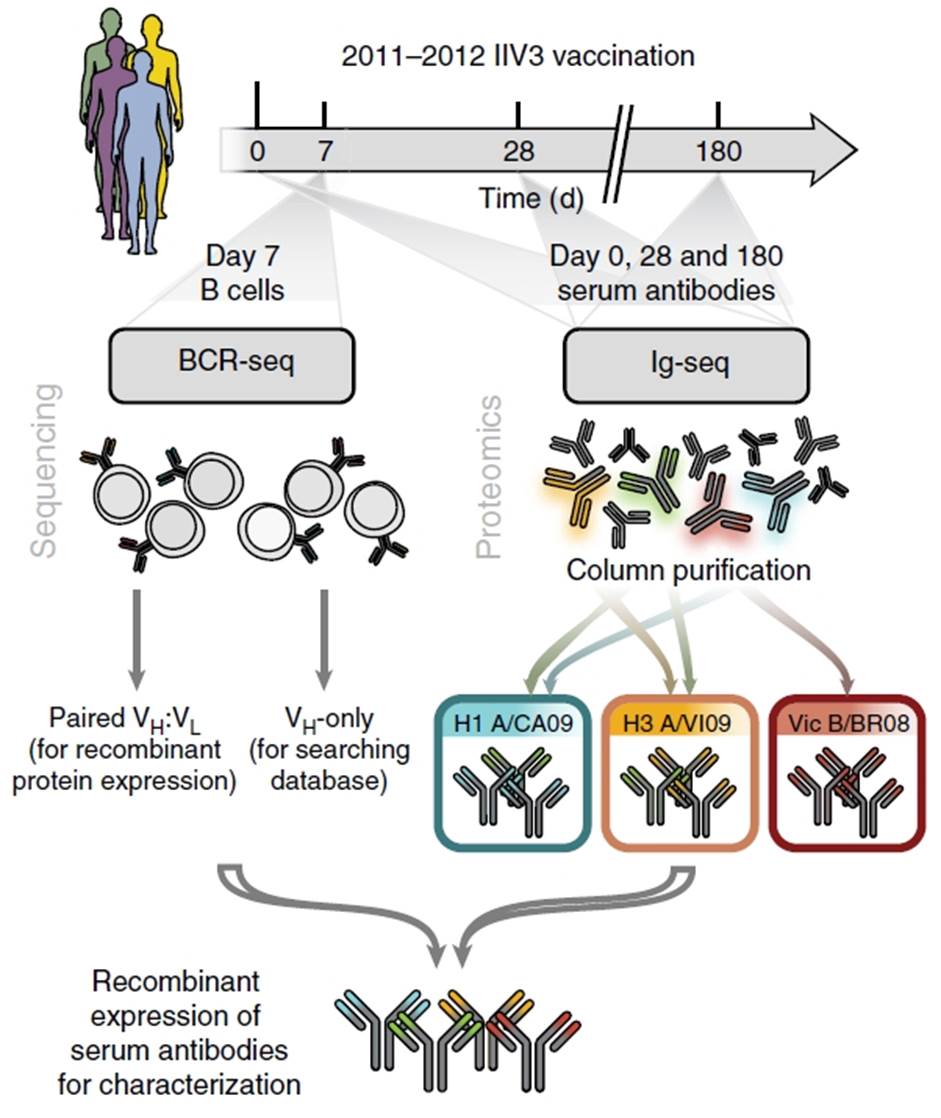
Figure 1. Study protocol for antibody repertoire analysis after influenza vaccination. BCR, B-cell receptor; Ig, immunoglobulin; VH, heavy chain, immunoglobulin variable region; VL, light chain, immunoglobulin variable region; Reproduced from Figure 1, panel a. Lee, J. et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med. doi:10.1038/nm.4224 (2016).
A second important observation came from analysis of H1/H3 cross-reactive antibodies detected by proteomics and affinity chromatography experiments. The authors generated custom databases containing the BCR sequences from peripheral B cells isolated after vaccination for each individual, and determined the full-length heavy and light chain sequences of seven of these cross-reactive antibodies to express them in a recombinant system for further study. Among the subset of H1/H3 cross-reactive antibodies analyzed, none had hemagglutinin inhibition activity and half showed no neutralization in pseudotyped particle-entry inhibition in in vitro assays. However, administration of three of these non-neutralizing antibodies protected mice from H1N1 infection, and two to H3N2, even at small doses. Additional negative-staining electron microscopy experiments led to the identification of a conserved HA epitope targeted by these antibodies.
A final insight of the paper should be highlighted: among the antibodies generated after trivalent vaccination (which includes an influenza B component) that recognize influenza B, several also reacted to a distinct influenza B component (B/Texas/6/2011). This means that the new quadrivalent IIV4 vaccine formulation might not offer significant additional protection over IIV3.
Highlighted paper:
Lee, J. et al. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med. doi:10.1038/nm.4224 (2016).
Further reading:
Georgiou, G. et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 32, 158–168 (2014).
Banner image credit: Getty Images/Ingram Publishing Thinkstock Images \ Ingram Publishing

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in