Journal Club | Pouring in metabolites without porins
Published in Microbiology

Authors: Members of the labs of Dr. Sarah Fortune and Dr. Eric Rubin*
Mycobacterium tuberculosis (Mtb), the etiological agent of tuberculosis, has a relatively impermeable cell envelope, which poses challenges in designing drugs that can effectively enter the cell. It also raises the question of how Mtb can obtain nutrients given that it seemingly lacks homologous mechanisms for import that have been described in other bacterial species. In recent work in Science, Wang et al. [1] find that members of a well-known but poorly understood PE/PPE protein family work in tandem to mediate selective uptake of diverse metabolites in Mtb, defining a novel mechanism for nutrient import in this important human pathogen.
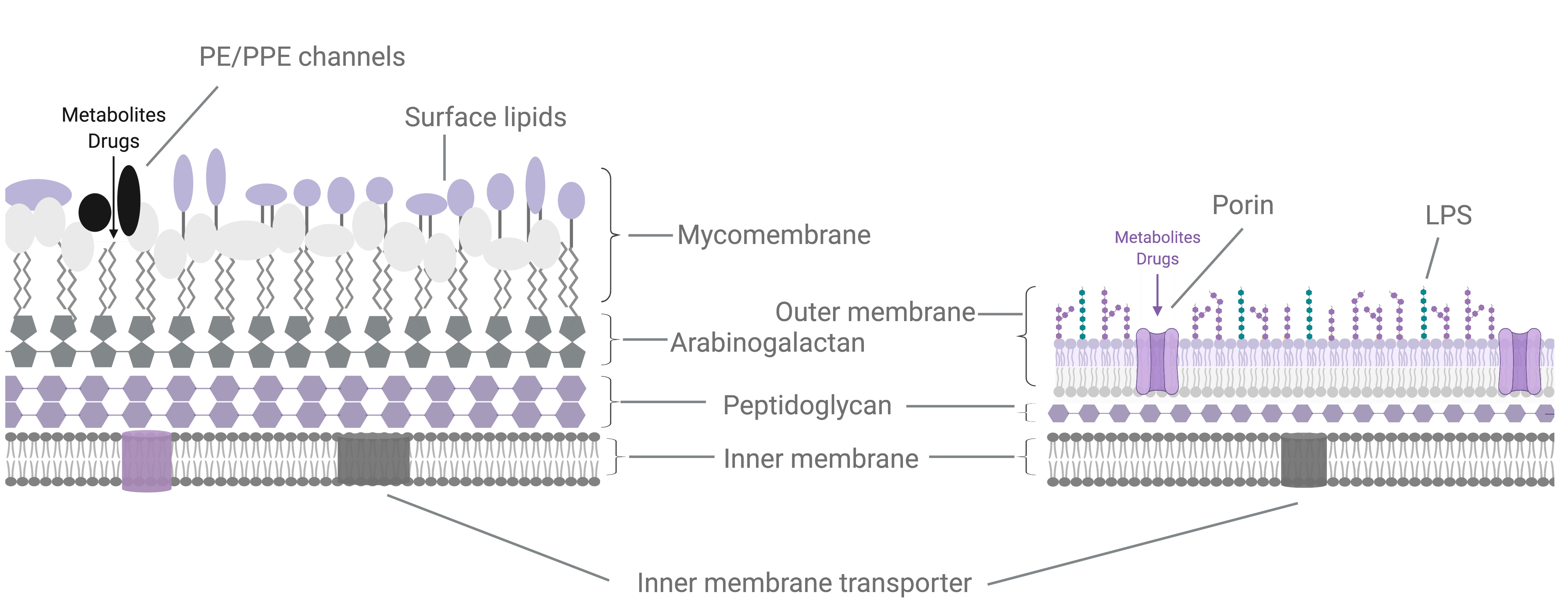
Mtb has a complex cell envelope with a lipid-rich outer membrane-like structure (the mycomembrane) that reduces the permeability of molecules and drugs [2]. The mycomembrane is composed of a variety of lipid species (Fig 1A), including phthiocerol dimycocerosate (PDIM), which is one of the most abundant lipids in the cell and has long been associated with Mtb virulence [3]. Recent studies have also shown that PDIM presents a permeability barrier and that certain M. marinum mutants have reduced growth unless PDIM biosynthesis is impaired [4].
Gram-negative bacteria, which have a similarly restrictive outer membrane, rely on a variety of proteins (porins) spanning the membrane to allow key molecules to enter the cell (Fig 1A). However, few porins have been demonstrated for Mtb. In their work, Wang et al. propose that different combinations of PE/PPE proteins serve a porin-like function to channel diverse molecules into Mtb. The PPE/PE family genes, so named based on conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) motifs in their N termini, are unique to mycobacteria and are particularly expanded in pathogenic slow-growing mycobacteria, with more than 160 PPE/PE genes annotated in Mtb [5]. Previous work has found that PE and PPE proteins are secreted as heterodimers by Type VII secretion systems and can be localized to cell envelope fractions. As such, PE and PPE genes have been proposed to serve diverse functions including immune evasion through antigen variation and the import of different nutrients [6], but to date the broader function of PE/PPE genes have remained largely unclear.
A
B
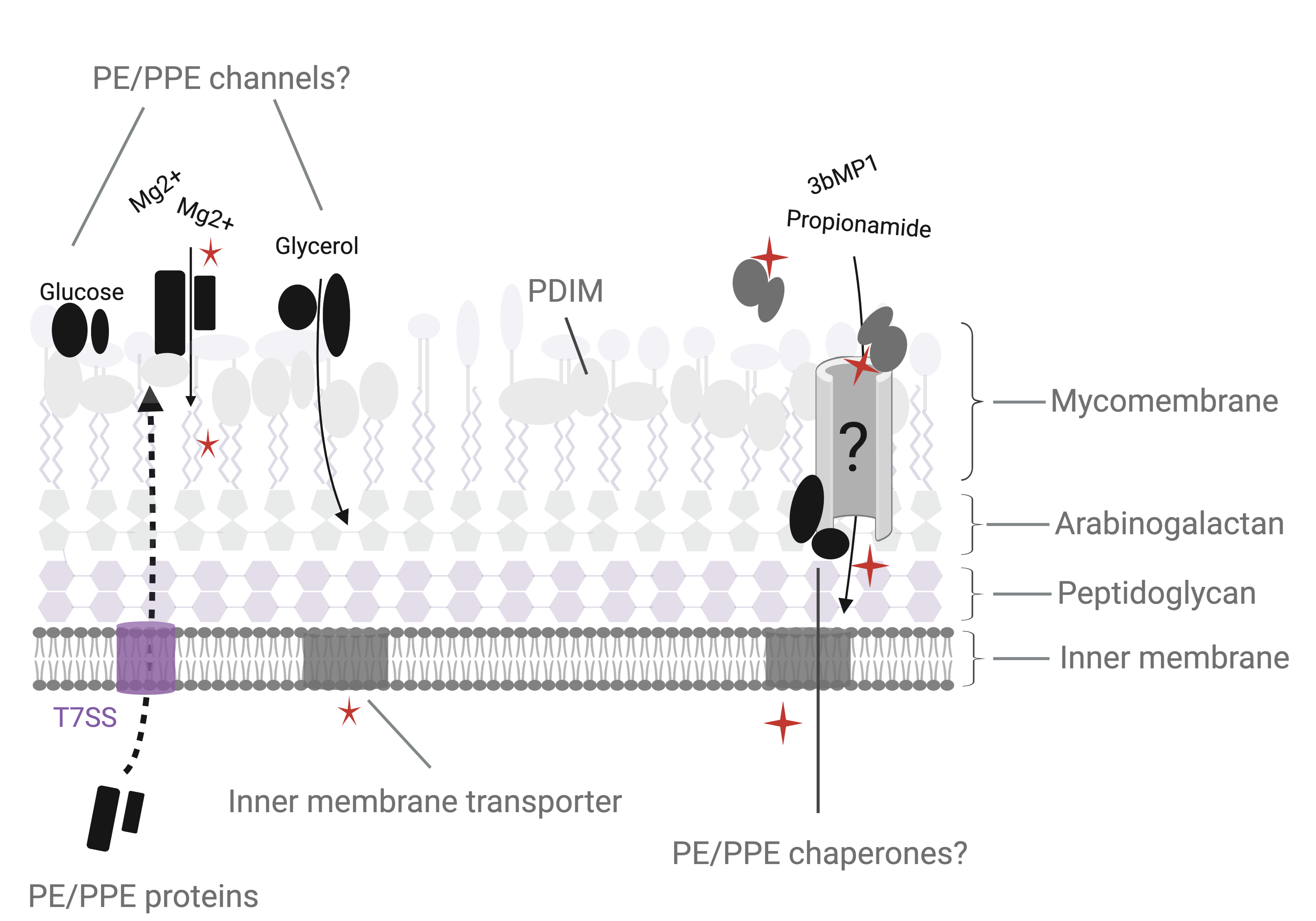
Figure 1. Metabolite and drug import in M. tuberculosis. (A) The M. tuberculosis cell envelope (left) is composed of an inner cell membrane, surrounded by 3 covalently linked layers of peptidoglycan, arabinogalactan and a lipid rich mycomembrane. The mycomembrane includes waxy mycolic acids that are covalently linked to the arabinogalactan layer as well as a variety of non-covalently embedded surface lipid species. The mycomembrane is proposed to function analogously to the lipopolysaccharide outer membrane found in Gram negative bacteria, which restricts diffusion of metabolites and drugs into the bacterial cell. (B) The authors propose that PE/PPE proteins, many of which are secreted by type VII secretion systems (T7SS) found on the inner membrane, can form porin-like channels that specifically allow metabolites and drugs through the PDIM-mediated permeability barrier within the mycomembrane. Alternative models may also exist for the PE/PPE proteins, including that PE/PPE pairs may be substrate binding proteins that deliver targets to other as yet undefined transporters, or that PE/PPE proteins may serve chaperone functions in aiding the biogenesis of porins.
In their study, Wang et al. first screened a compound library and discovered a compound (3bMP1) with bactericidal activity against Mtb. When they selected for spontaneous resistance mutations to 3bMP1, they surprisingly found genomic changes mapping to the ppe51 gene. Given chemical similarity between 3bMP1 and the metabolite propionamide, they next showed that 3bMP1 resistant mutants also lost the ability to grow on propionamide as the sole carbon source. Interestingly, the resistant mutant was also defective for growth on and uptake of additional carbon substrates including glycerol and glucose but not of others like hexanamide.
It is particularly laudable that when the authors deleted ppe51 in Mtb and found that the strain did not reproduce their 3bMP1 resistance phenotype, they pursued why and discovered that their mutant harbored a second site mutation that abrogated production of PDIM. Constructing more Mtb mutants subsequently confirmed that ppe51 was required for 3bMP1 sensitivity and import of glucose and glycerol and that loss of PDIM can indeed reverse this requirement. Additionally, biochemical fractionation confirmed that PPE51 is enriched in the Mtb mycomembrane fraction.
The authors next undertook a transposon screen to identify additional genes involved in 3bMP1 resistance and found several mutations in ppe51 and pe19. Deletion of pe19 recapitulated the Δppe51 drug and growth phenotypes and, as expected, these were suppressed by PDIM deletion. As PPE and PE proteins are thought to work in complex, the authors biochemically showed that PPE51 and PE19 interact in Mtb. Together, these data lead the authors to propose a model by which PPE51 and PE19 may form porin-like channels that allow specific metabolites to cross an impermeable PDIM barrier (Figure 1B).
Finally, Wang et al. tested additional PE/PPE pairs for metabolite-dependent phenotypes. Through targeted deletion or CRISPRi-based knockdowns they show that PE20/PPE33 and PE19/PPE25 pairs were required for Mtb growth during magnesium and phosphate limitation, respectively. While further experiments to measure substrate import in these mutants and direct interactions between these PE/PPE pairs would be important to confirm that these proteins may be working in complex as proposed for PPE51 and PE19, these experiments suggest that PE/PPE family proteins may operate combinatorially to import a variety metabolites and drugs in Mtb (Fig 1B).
This work provides a compelling finding that PE/PPE proteins play a role in substrate-specific import in Mtb. While the authors propose that these pairs may operate as channels, other models may also be plausible, including PE/PPE pairs acting as specific substrate binding proteins that deliver molecules to an as yet undefined transporter or PE/PPE pairs being in involved in the function or biogenesis of other transporters (Fig 1B). Future biochemical and structural characterization of PPE51-PE19 will help define the ability of this complex to interact with substrates and form higher order structures reminiscent of membrane transport systems (e.g. a recent study defined the structure of a Mtb ABC transporter of certain hydrophilic compounds [7]).
Previous work has shown that expression of PE and PPE genes is regulated during infection and by different growth conditions [5,6], and Wang et al. show that PE20 and PPE33 mutants were most defective for growth in magnesium limitation in mildly acidic pH. Given the large PE/PPE repertoire in Mtb, one could envision that subsets of genes are expressed and utilized by Mtb to adapt to diverse host environments offering different stressors and nutrients. These functions could also be a feature of other type VII secreted proteins given that some, like EspA/C, have structural homology to PE/PPE complexes and a mutant in EspA had increased susceptibility to cell wall stress [8].
It has been proposed that impermeability is a feature associated with the evolution of virulence across mycobacterial species [9] and it would be interesting to assess emergence of PDIM in mycobacteria relative to the expansion of PE/PPE protein families, which may suggest that acquisition of pathogenicity drove compensatory evolution of diverse transport systems to mitigate the effects of PDIM-mediated nutrient restriction. This relationship could also be useful in the context of drug development—inhibitors of PE/PPE could be less likely to select for resistant strains with competitive fitness, as compensatory loss of PDIM to increase permeability would lead to attenuation in virulence.
In conclusion, Wang et al. have established an important framework for understanding how Mtb has solved the critical problem of selectively obtaining nutrients. They have also advanced our understanding behind the role of the enigmatic PE/PPE family proteins, an impressive feat given these genes and proteins are often left out of analyses due to their highly repetitive nature. Isn’t it incumbent upon us to do the same?
*Acknowledgments:
This was a collaborative project between members of Dr. Sarah Fortune's and Dr. Eric Rubin's labs. Many thanks to all who participated in the discussion of the work and the following people for helping write this piece and design figures (images made on https://biorender.com): Thibualt Barbier, Allison Carey, Michael Chao, Nicole Howard, Qingyun Liu, and Francesca Tomasi.
References:
1. Wang Q et al. Science 367, 1147-1151 (2020)
2. Dulberger, CL, Rubin EJ, Boutte CC. Nature Reviews Microbiology 18, 47-59 (2020)
3. Arbues A et al. Front Cell Infect Microbiol 4, 173 (2014)
4. Ates LS, et al. PLoS Genet 11,e1005190 (2015)
5. Fishbein S et al. Mol Micro 96, 901-16 (2015)
6. Ates LS. Mol Micro 113, 4-21 (2020)
7. Rempel C et al. Nature - doi: 10.1038/s41586-020-2072-8 (2020).
8. Garces A et al., PLoS Pathog 24, e1000957 (2010)
9. Jankute M et al. Sci Rep 7,1315 (2017





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in