Trypsin is the most commonly used protease for mass spectrometry–based proteomics experiments, because of its well defined specificity (it results in peptides with either lysine or arginine at the C-termini).
With an increasing focus on answering the question “How many copies of this protein are present per cell?”, it becomes important to make sure that the protease digestion does not result in more- or fewer-than-expected peptides for any given protein.
Is trypsin always the best protease for such proteomics experiments?
It turns out that it would be a good idea to try other proteases as well.
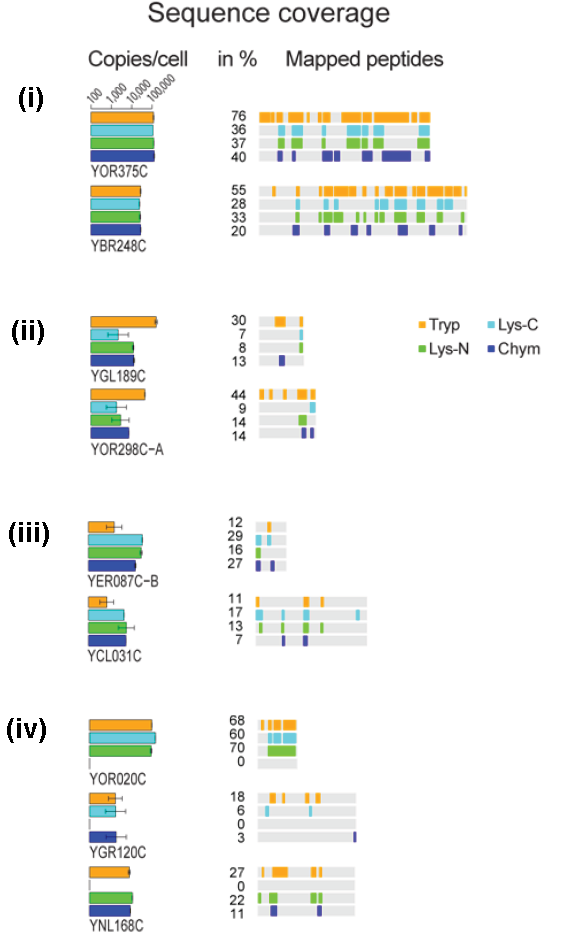
In a letter to Nature Methods, Peng et al. describe results from an experiment where they treated aliquots from a yeast lysate with different proteases (trypsin, Lys-C, Lys-N and chymotrypsin). They then performed strong cation exchange chromatography followed by LC–MS/MS and generated proteomics data sets based on both spectral counts and ion volume. While there was a good correlation between the results obtained from technical replicates, correlation between samples obtained using different proteases was less good, an effect which was especially pronounced with chymotrypsin, which has a different cleavage specificity.
The authors found that while amount of some proteins was similar no matter what protease was used (e.g. those shown in (i) below), there were others where using trypsin seemingly over-estimated the copy number (ii) OR underestimated the copy number (iii), and there were proteins which would not have been seen if only one of the proteases had been used (iv).
(the Supplementary information provides a lot of information regarding the methods used, and includes a more comprehensive set of results).
Mao Peng, Nadia Taouatas, Salvatore Cappadona, Bas van Breukelen, Shabaz Mohammed, Arjen Scholten & Albert J R Heck. Protease bias in absolute protein quantitation. Nature Methods 9, 524-525 (2012)
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in