Machine learning is revolutionizing the search for new HIV integrase inhibitors.
Published in Computational Sciences and General & Internal Medicine
Explore the Research
20250719-392359-vi423u.pdf?utm_campaign=related_content&utm_source=HEALTH&utm_medium=communities
Review began 06/10/2025
Overview of HIV research with Machine learning
HIV/AIDS continues to pose a significant public health challenge, with an estimated 39 million people affected worldwide. Of these, approximately 25.6 million reside in Sub-Saharan Africa. According to the World Health Organization (WHO) and UNAIDS, East and Southern Africa bear more than half of the global HIV burden [1]. Machine learning (ML) technologies are playing an expanding role in HIV research, particularly in accelerating drug discovery through the identification of novel therapeutic targets and optimization of candidate compounds. In addition to advancing understanding of viral mechanisms, ML contributes to innovation in diagnostics, treatment personalization, and prevention strategies. Despite challenges such as data limitations and model transparency, ML holds substantial promise in enhancing HIV-1 drug development and supporting global health efforts [2].
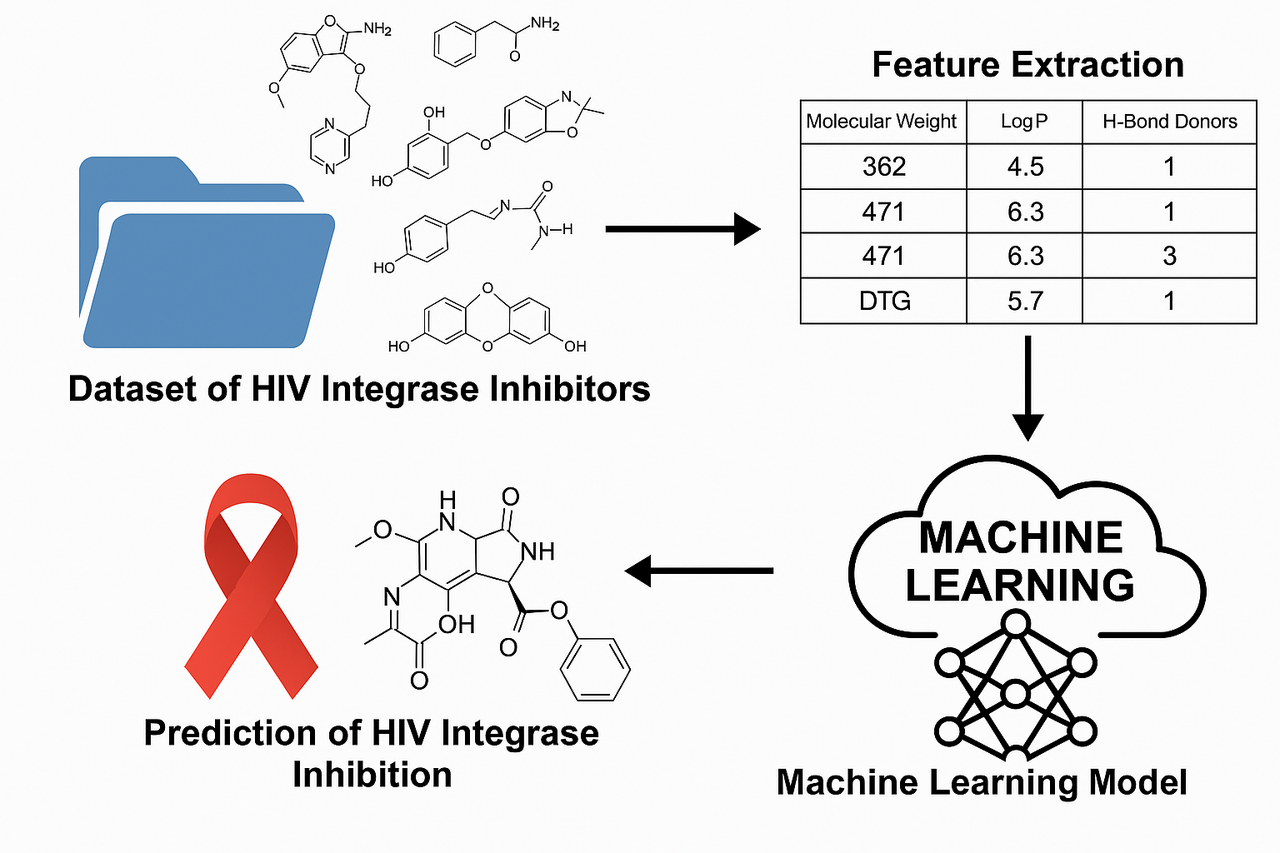
A pharmacist from Zimbabwe published a paper in Cureus journal about “Development and Validation of a Machine Learning Model for Identifying Novel HIV Integrase Inhibitors” , This study uses machine learning (random forest and logistic regression) to predict HIV integrase inhibitors from a curated dataset. It identifies key molecular descriptors driving inhibition and achieves strong predictive performance (AUC-ROC: 0.886). The work demonstrates ML's potential in accelerating HIV drug discovery and proposes future development of an accessible prediction platform. This PDF for this paper is downloadable on (Cureus 17(6): e86326. doi:10.7759/cureus.86326).
Utilizing a curated dataset from the ChEMBL database, the study extracted molecular descriptors from known integrase inhibitors, using cheminformatics tools like RDKit. Preprocessing steps included standardizing SMILES notations, converting IC50 to pIC50 for better model performance, and removing incomplete or implausible entries. Descriptors such as molecular weight, LogP (lipophilicity), hydrogen bond donors/acceptors, topological polar surface area (TPSA), and rotatable bonds were computed and analyzed for feature selection.
The study implemented two machine learning models: logistic regression and random forest, using a binary classification approach to predict inhibitory potential. A threshold pIC50 value of 5 was used to classify compounds as inhibitors or non-inhibitors. Recursive feature elimination helped identify the most predictive features, and hyperparameter tuning optimized model performance.
Evaluation metrics—accuracy, precision, recall, F1-score, and area under the ROC curve (AUC-ROC)—revealed that the random forest model significantly outperformed logistic regression. The random forest model achieved an AUC-ROC of 0.886, accuracy of 0.816, and precision of 0.792, compared to logistic regression's AUC-ROC of 0.595 and accuracy of 0.580. Feature importance analysis highlighted TPSA, LogP, and molecular weight as the most influential descriptors in predicting inhibitory activity. The strong correlation between several descriptors (r > 0.90) suggested multicollinearity, particularly affecting logistic regression performance.
One of the most active compounds identified had a pIC50 of 9.34 and demonstrated a favorable balance between lipophilicity and hydrophilicity, with promising structural properties for biological interaction. The ROC curve further illustrated the random forest model’s superior ability to discriminate between inhibitors and non-inhibitors.
The study underscores the value of ensemble learning methods like random forest in cheminformatics, especially when handling nonlinear feature interactions. Compared to similar studies, this work focused on descriptor-based modeling and internal validation, offering a clear pathway for integrating ML into early-phase drug discovery.
Despite its strengths, limitations included dataset size, lack of external validation, and potential overfitting. The binary classification approach may have oversimplified the continuous nature of bioactivity data. Future work should incorporate external datasets, explore deep learning models, and validate predicted compounds experimentally.
Potential Uses for this technique
- High-Throughput Virtual Screening of Large Compound Libraries
The model can be used to rapidly screen millions of compounds from commercial or public chemical libraries (e.g., ZINC, ChemBridge) to identify promising HIV integrase inhibitors. This significantly reduces the cost and time required for laboratory-based screening and helps narrow down candidates for in vitro or in vivo testing.
- Integration into Drug Discovery Platforms (e.g., Streamlit-based Tools)
By deploying the model as a user-friendly web application using tools like Streamlit, researchers without coding skills could input chemical structures (via SMILES) and instantly receive predictions about integrase inhibition potential. This can democratize access to ML-powered drug discovery tools, particularly in low-resource settings or academic institutions.
2. Expansion to Multi-Target HIV Drug Discovery Models
The model’s architecture can be adapted to predict inhibitors for other HIV targets (e.g., reverse transcriptase, protease, or capsid proteins). By expanding descriptor datasets and training on multi-target profiles, it could support the development of dual- or multi-action antiretroviral agents, addressing resistance and improving therapeutic efficacy.
References
- Jaiteh M, Phalane E, Shiferaw YA, Phaswana-Mafuya RN. The application of machine learning algorithms to predict HIV testing in Repeated Adult Population–Based Surveys in South Africa: Protocol for a Multiwave Cross-Sectional Analysis. JMIR Research Protocols [Internet]. 2025 Jan 27;14:e59916. Available from: https://www.researchprotocols.org/2025/1/e59916
- Jin R, Zhang L. AI applications in HIV research: advances and future directions. Frontiers in Microbiology [Internet]. 2025 Feb 20;16. Available from: https://doi.org/10.3389/fmicb.2025.1541942
- Mukuhlani BT. Development and validation of a machine learning model for identifying novel HIV integrase inhibitors. Cureus [Internet]. 2025 Jun 18; Available from: https://doi.org/10.7759/cureus.86326
Follow the Topic
-
Cureus Journal of Computer Science

Cureus Journal of Computer Science offers a platform for credible research and innovative perspectives, while promoting inclusive and diverse article formats in all areas of computer science.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in