Making cells bigger to see what was previously invisible
Published in Protocols & Methods and Cell & Molecular Biology
Visualizing intracellular signal flow in living cells by creating functional giant cells
Understanding how signals propagate inside a living cell has long been a fundamental challenge in cell biology. While chemical messengers such as cyclic AMP (cAMP) and calcium ions (Ca²⁺) orchestrate cell polarity, migration, and collective behaviors within seconds, directly observing how these signals move through the interior of a single cell has remained technically difficult. The reason is simple: cells are small, and diffusion-driven signals spread too quickly to resolve directionality within the spatial and temporal limits of conventional microscopy.
In our recent study, published in Communications Biology, we addressed this challenge by taking a deliberately unconventional approach: instead of trying to improve microscopy further, we made the cell itself larger.
A simple idea: inhibit division, not function
Using the social amoeba Dictyostelium discoideum as a model organism, we selectively inhibited cell division while preserving cellular viability and physiological function. This allowed individual cells to grow to more than 100 times their normal volume without genetic manipulation. Importantly, these “giant cells” retained key biological behaviors, including chemotaxis toward cAMP, front–rear polarity formation, and dynamic cytoskeletal activity.
Unlike expansion microscopy or chemical swelling techniques, which require fixed samples, our method produces living enlarged cells. This effectively expands the intracellular landscape, pushing the optical resolution of standard fluorescence microscopy beyond its usual limits and allowing us to track fast, diffusive signals in real time.
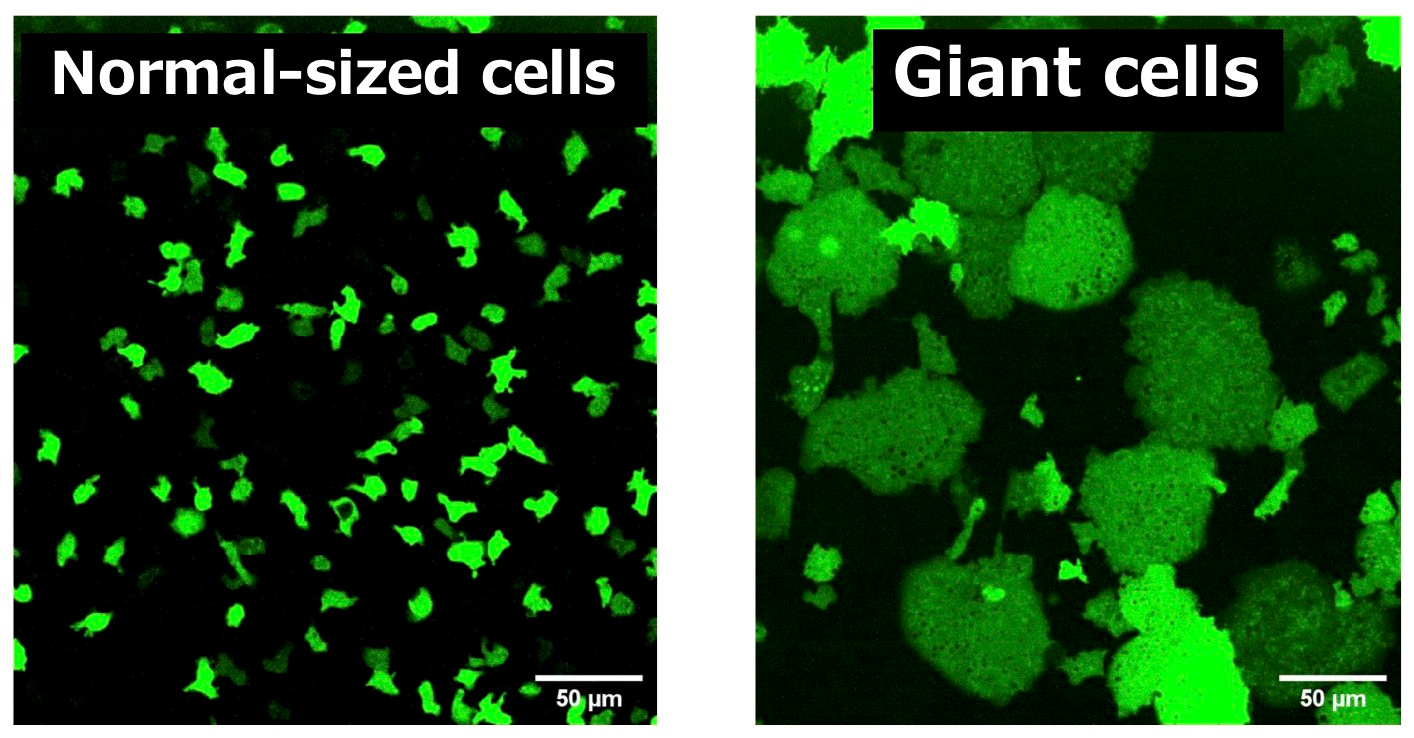
Directly observing front–rear signal propagation
By combining giant cells with high-sensitivity fluorescent biosensors, we were able to visualize intracellular signaling in ways that were previously impossible.
When monitoring cAMP dynamics using the Flamindo2 sensor, we observed a clear directional asymmetry: cAMP levels rose first at the cell front (the side facing external stimulation), while signal decay consistently initiated at the rear. This front-to-rear propagation pattern had been hypothesized for decades but had never been directly visualized within a single living cell.
Interestingly, the posterior region of the giant cells contained dense clusters of vesicles. Our observations suggest that these vesicles may contribute to the rapid local clearance or buffering of cAMP, shaping intracellular gradients that support persistent polarity.
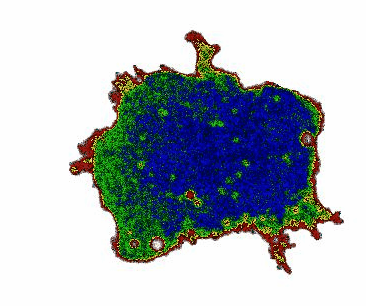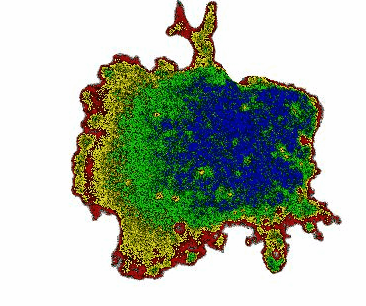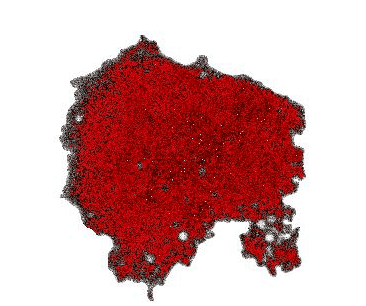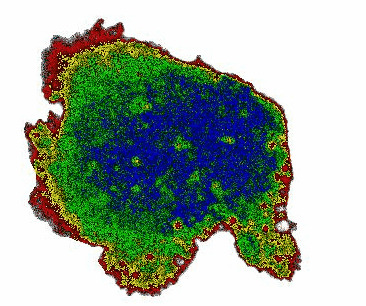
Beyond Dictyostelium: a general platform for cell biology
Our work demonstrates that functional giant cells can serve as an expanded, living model of single-cell information processing. By physically enlarging the cell rather than chemically fixing or computationally inferring dynamics, we gain direct access to the “real-time map” of intracellular signaling.
We anticipate that this approach will be applicable to other signaling molecules and, potentially, to mammalian or plant cells. More broadly, it offers a new experimental platform for addressing a fundamental question in biology: how does a cell integrate spatial information to make decisions?
By making cells bigger, we can finally see what was always there—but too small to observe.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in