Manage life science research data with pISA-tree
Published in Research Data

In life science research projects, large amounts of diverse data are generated, thus managing them in a findable, accessible, interoperable and reusable (FAIR) manner is a big challenge. To fast-track discoveries, wider implementation of open data management following FAIR principles is imperative.
As simple as possible, as detailed as necessary
Although researchers in life sciences are generally fond of the idea of open science and FAIR data, not many are keen on spending their working days organising data files and filling endless metadata tables. Especially frustrating is when, for example, you have to store data for the same samples in different data type-specific systems or when leaving a field empty in a metadata form is not allowed. Scientists want to have their data organized, however usually in a manner that is as simple as possible and as detailed as necessary. The simplest way of organising data files in computers is within a directory structure. It is transparent, hierarchical, and easy to navigate.

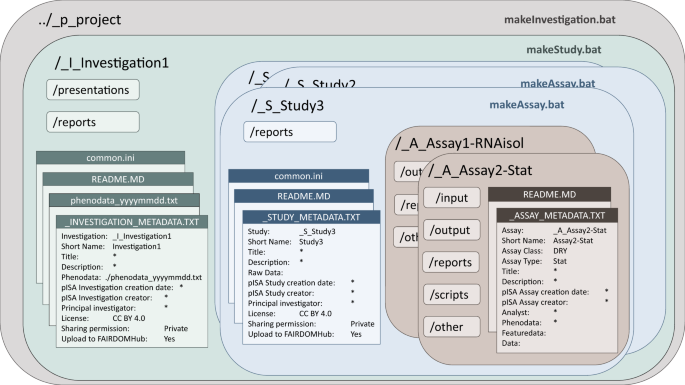
Why we developed pISA-tree?
In our lab, researchers were already organising their project data in directories, the only problem was that it was not organised in a way that colleagues could find and make sense of it. This was simply because everyone had his own way of organising the files. In addition, metadata were scattered, hidden in paper lab diaries and not collected systematically. That is why pISA-tree was built jointly by computer scientists and biologists, to capture all requirements of FAIR guiding principles and metadata standards, but, on the other hand, also make it in line with the thinking of biologists. pISA-tree is thus prepared in a way to facilitate metadata generation with each experiment, when the samples are collected and data generated. This allows the users to have all data linked and FAIRly organised from the start of the project. Consequently, the stress of organising the data when preparing a publication is considerably reduced. pISA-tree was tested in several international and national projects and was appreciated by the users of both sides, the producers of data and data analysts. It is also experiment type agnostic, we tested it for handling confocal imaging, NGS, metabolomics, metagenomics, whole genome sequencing and bioinformatics analysis as well as model simulations. We describe pISA-tree in a newly published Scientific Data article.
How can you use pISA-tree and contribute?
Download pISA-tree from GitHub and read the instructions. You can also create your custom metadata templates and share them with the community by uploading them to https://github.com/NIB-SI/pISA-tree-assay-types.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Genomics in freshwater and marine science
Publishing Model: Open Access
Deadline: Jul 23, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in