Mechanisms of Pseudomonas aeruginosa resistance to type VI secretion system attacks
Published in Microbiology
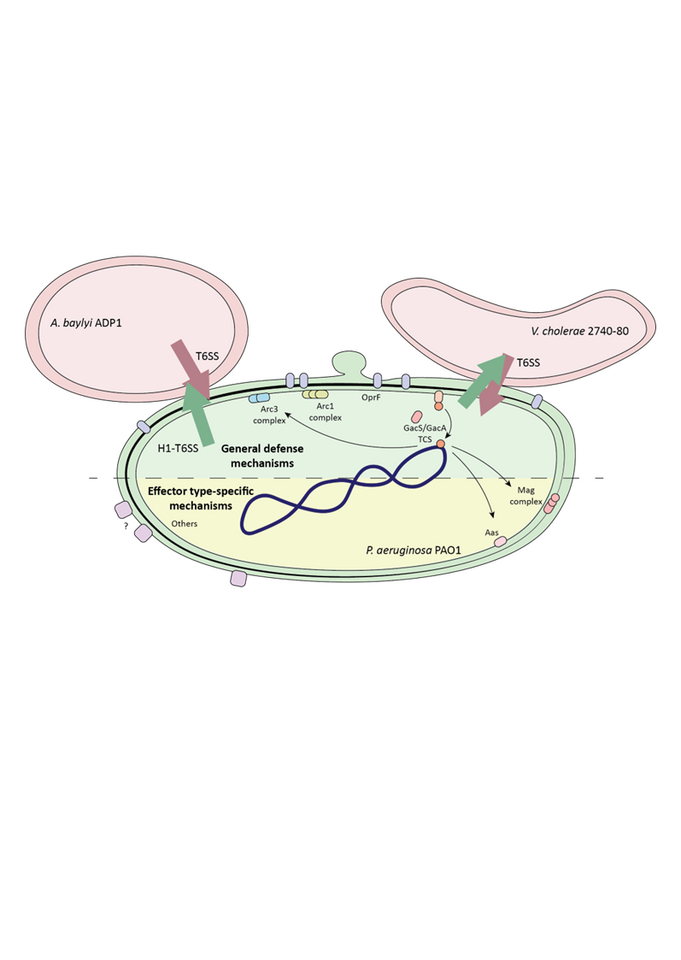
In the environment, bacteria grow mostly on surfaces, where they frequently encounter other species that compete with them for scarce resources. Over time, certain bacteria have developed sophisticated molecular machineries to outcompete, and even attack, their neighbors. One of these machineries is the Type VI Secretion System (T6SS), a molecular nanoweapon that assembles inside the attacker bacterium and shoots a spear-like structure loaded with toxic effector proteins into neighboring bacteria. These effector proteins have different enzymatic activities and are often able to kill the prey cell within minutes.
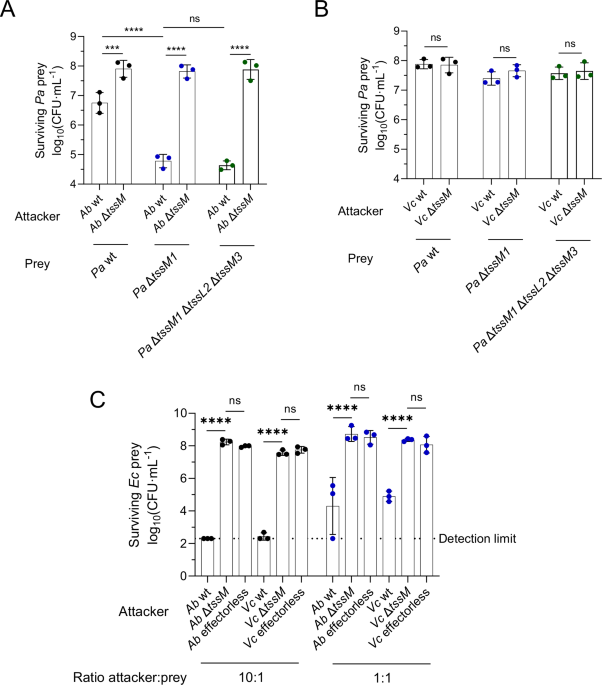
Some bacteria have very active T6SSs that constantly shoot effectors into their environment, regardless of whether competitors are around, a strategy that may represent an energetic burden for the cell under conditions of scarce nutrients. However, other bacteria have developed more elaborate strategies to regulate the assembly and firing of their T6SS. One striking example is the T6SS of Pseudomonas aeruginosa, an environmental bacterium and opportunistic human pathogen. P. aeruginosa first senses membrane perturbations inflicted by competitors (through their own T6SS or other weapons), and in response triggers a T6SS assembly directed specifically towards the origin of the damage. In this way, P. aeruginosa can direct its efforts towards killing the competitor bacteria without constantly firing its own T6SS. At the start of this study, we reasoned that this must mean that P. aeruginosa is able to survive the injection of toxic effectors from neighboring bacteria, at least long enough to mount a retaliatory response. Indeed, our data indicated that P. aeruginosa is much more resistant to T6SS attacks than other bacteria, suggesting the existence of uncharacterized T6SS resistance mechanisms.
In parallel, our collaborators in the lab of Urs Jenal developed a CRISPRi library that allows the screening of a collection of P. aeruginosa strains with decreased expression of every single gene, enabling the determination of genes that are important for survival in a specific environmental condition. We decided to apply this library to an interbacterial competition setting, where P. aeruginosa faces competitors with or without an active T6SS. This allowed us to pinpoint genes that are important for survival to T6SS attacks, and specifically against certain effector classes like lipases or peptidoglycan-targeting effectors. We could then modify the genome of P. aeruginosa to remove these genes and further study their impact on survival of P. aeruginosa against T6SS attacks. Among other genes, we found a group of genes, a so-called operon, that are linked to each other and often work together to carry out a common task. We found that this operon, called the mag operon, is distantly related to eukaryotic immune systems and protects P. aeruginosa against toxins that degrade its peptidoglycan, and maybe others. In the video below you can see a small timelapse showing mutant cells of P. aeruginosa dying faster than the original strain due to T6SS attacks.
Video 1. Timelapse imaging of Acinetobacter baylyi (T6SS sheath protein labeled in green) competing with P. aeruginosa (in black). On the left panel there is P. aeruginosa WT, and on the right panel a P. aeruginosa strain lacking gacA (right panel). In cyan are dying P. aeruginosa cells, that become permeable as a consequence of T6SS attacks from A. baylyi.
In recent years, it has become apparent that bacteria also have sophisticated immune systems, not unlike those of eukaryotic cells. These immune systems have primarily been studied in the context of bacterial defense against bacteriophages that act as parasites of these bacteria. Our study also suggests that some bacteria have developed an immune system against T6SS toxins that requires further study, and that there may be an ongoing evolutionary arms race to obtain new weapons that give bacteria a competitive advantage in their environment; as well as to obtain new defense mechanisms against these weapons.
This raised the question, why not accumulate as many of these defense mechanisms as possible? The more defense mechanisms the better, right? In our study, we also looked at how these T6SS defense mechanisms impact the susceptibility of P. aeruginosa to antibiotics. Surprisingly, we found that P. aeruginosa that contains some of these resistance mechanisms is actually more sensitive to some families of antibiotics, indicating that there is a trade-off to the accumulation of these protective genes, and that they may not always provide an advantage.
But, what is the right balance? We need to do more research to understand why these T6SS resistance mechanisms can lead to antibiotic susceptibility, and in which contexts this may be important. Also, what happens during human infection? How does the presence of these T6SS resistance mechanisms impact competition with other co-colonizing bacteria, and how do they impact survival of P. aeruginosa after antibiotic treatment? Our findings may pave the way to understanding the interplay between P. aeruginosa, its competitors and antibiotics within a human host.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in