New Collection on The Pan-Cancer Project
Published in Cancer

Explore the Research

Pan-Cancer Analysis of Whole Genomes
Cancer is a disease of the genome, caused by a cell's acquisition of somatic mutations in key cancer genes.
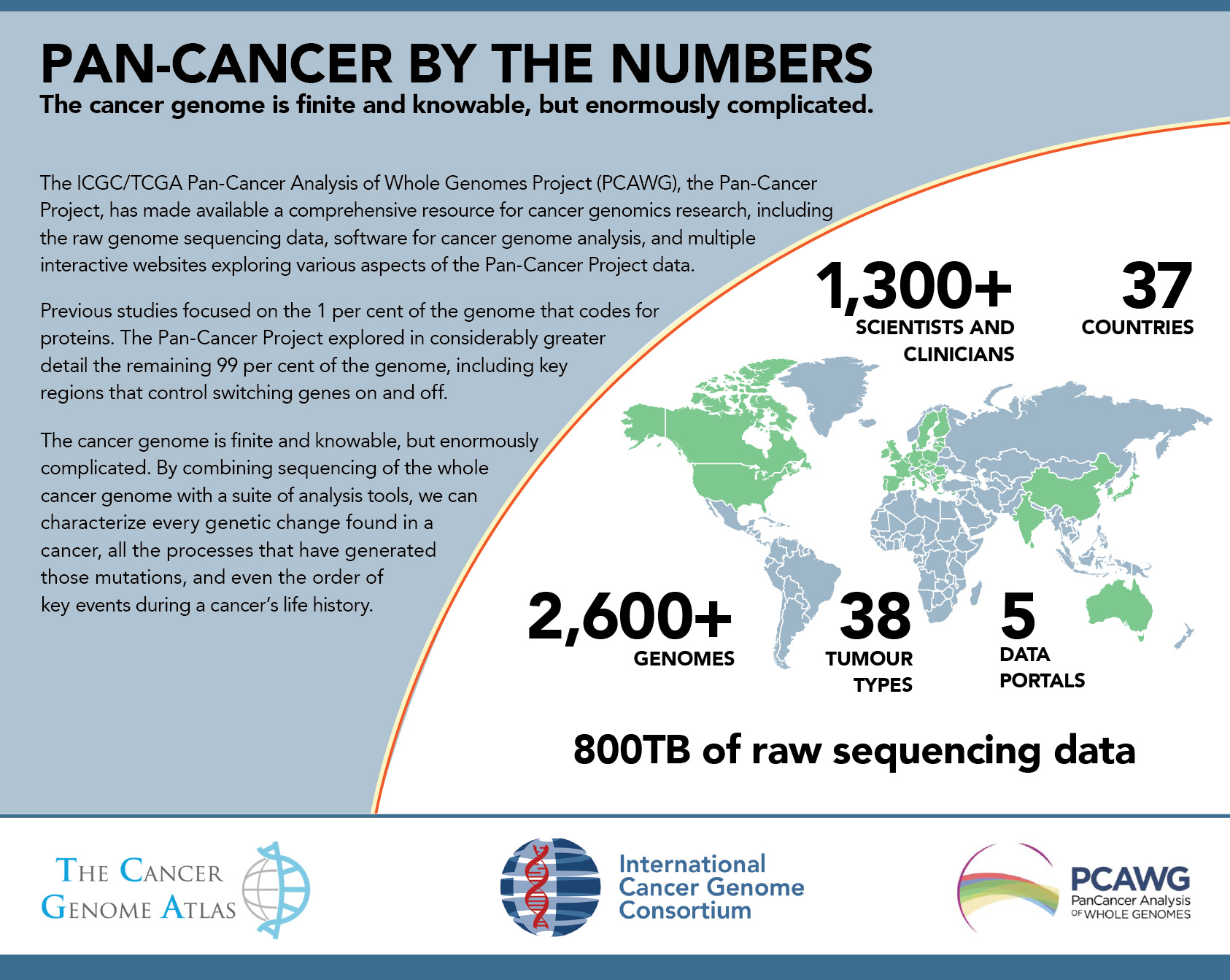
Building on previous work of The Cancer Genome Atlas (TCGA), which examined cancer coding regions, The Pan-Cancer Analysis of Whole Genome (PCAWG) Project, known as the Pan-Cancer Project, is an international collaboration of more than a thousand researchers and clinicians to identify common patterns of mutation in more than 2,600 whole cancer genomes in 38 tumour types from the International Cancer Genome Consortium (ICGC). It’s the most comprehensive analysis of cancer genomes to date.
The Pan-Cancer Project explored the nature and consequences of DNA variations in cancer, across the entire genome, from both protein-coding genes and from areas of DNA that do not code for proteins. The initiative revealed the extensive role played by large-scale structural mutations in cancer, identified previously-unknown cancer-related mutations in gene regulatory regions, inferred tumor evolution across multiple cancer types, illuminated the interactions between somatic mutations and the transcriptome, and studied the role of germline genetic variants in modulating mutational processes. This collection comprises papers describing the core set of analyses conducted by the Pan-Cancer Consortium, and showcases data, tools, and other resources useful for those who seek to further explore this legacy data set.
The Pan-Cancer data is available for the broader research community to use, enabling a wider and deeper exploration of the cancer genome, by making sequencing data on genomes’ non-coding regions available and providing tools to examine this data.
Understanding the genomic changes in many forms of cancer worldwide will enable further research into the causes, prevention, diagnosis and treatment of cancers. Cancer genomes are complex, and much more data, potentially in thousands to tens of thousands of patients per tumour type, are needed to fully understand them, which is why shared data and resources like the Pan-Cancer Project are so important.
Collection: Pan-Cancer Analysis of Whole Genomes
Poster image: the Pan Cancer Project team (Stuart Lawler, OICR).

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in