
Back in September Springer Nature and protocols.io announced a new partnership which, through looking at the use of protocols, is set to explore the developments in open research practices. Having easy and open access to protocols and methods facilitates discussion, revision, optimization, and reuse and as such sits at the heart of reproducibility in open science. Therefore, clear understanding of their use, researcher need, and implementation is critical to ensure that research results are replicable and that research time and money is not wasted.
As a leading publisher of protocols, Springer Nature has a commitment to the community to not only support researcher needs but to best understand how they wish to use and re-use such material, adapting to technological and methodological changes and requirements to enable this. But what does this look like in practice for a researcher and fundamentally what does the drive around protocol use mean more practically for research and the sector? As a champion of the benefits of sharing dynamic protocols, research Microbiologist Ruth.E. Timme discusses further.
Ruth is an author of an OA chapter with Springer Nature that she had placed onto protocols.io prior to the Springer Nature-protocols.io partnership. You can see how the book chapter is dynamically represented as a collection of protocols on protocols.io here.
Benefits of the protocols.io platform for a public health laboratory network.
Just as GenomeTrakr, an open-access genomic epidemiology database has helped support great advances in identifying the sources of foodborne contamination events, we expect open-access repositories of protocols to help researchers follow the most up-to-date guidance and Best Practices for generating and submitting genomic epidemiology data worldwide. This sharing is especially important during a global pandemic, where new methods need to be communicated globally and updated quickly.
Where this started
In 2019, my colleagues and I published a chapter in Methods in Molecular Biology: this was a set of protocols for sharing genomic epidemiology data from foodborne pathogens in a public repository. As I was writing that chapter, I was concerned about the evolving nature of dry-lab protocols. These methods were not supposed to be set in stone, but to change as technology and understanding developed over time. How was I going to communicate routine updates to laboratories across the world, so they could follow the most current version of these protocols?
Then I attended a workshop where I learned about the protocols.io open access platform, which seemed to be a great solution to my problem. I immediately created updated versions on the protocols.io platform and referenced them in a new publication, laying out these Best Practices for laboratories to follow when contributing to our open genomic epidemiology effort. Now, when refinements are made or new tools become available, participants can keep up to date with one another, while still being able to see how the earlier work had been performed.
What did we do before?
Prior to protocols.io, we would create protocols in an MS Word document, then create a PDF and upload to a private shared space accessible only to our own laboratories. Any updates to these protocols would be communicated via email or during monthly video calls. Shared protocols on protocols.io let us do science, test out methods, and share our customisations without having to manually track, organise, and distribute individual protocol PDF files and their many versions. Working hours can be spent more productively: we can now create new protocols within the platform, then simply “publish” to make them publicly available.
Beyond this, collaborators in the GenomeTrakr project have access to DRAFT protocols inside our dedicated GenomeTrakr workspace and can test these protocols out in their own labs prior to formal publication. Their experimentation gives us valuable feedback prior to sharing the protocols more widely. Members of the GenomeTrakr workspace can also receive immediate notifications when comments and updates are made on protocols they are following, enabling our global community to stay up-to-date without us having to track and manually disseminate all these details.
Finally, knowing which version of a protocol was used for a given experiment is now simple and clear, thanks to built-in DOI tracking.
The role of open access (OA)
As shown in the diagram below, the cornerstone for these successful global, collaborative projects is open access: open access protocols (the protocols.io GenomeTrakr workspace), open access database (NCBI), and open access analysis tools (GalaxyTrakr).
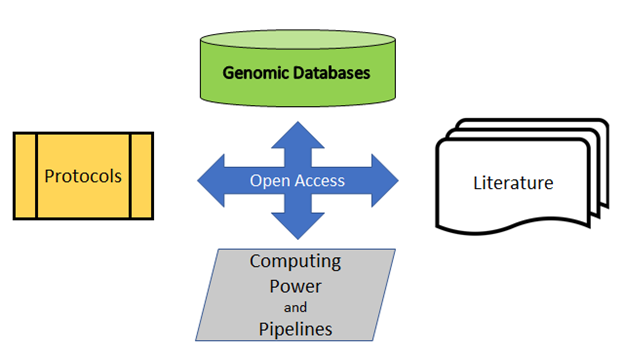
Open access to open resources allows research to thrive. Open resources also support the goals of the FAIR Guiding Principles for scientific data management and stewardship: data and metadata should be Findable, Accessible, Interoperable, and Reusable.
What have these accessible, adaptable and replicable resources accomplished so far?
We have used protocols.io as a resource for our collaborators across the global GenomeTrakr community. The platform allows users to comment on specific steps, enabling us to identify and track steps that need clarification or updating. Keeping our protocols current is also easier, since version control is built into the platform. We can easily publish new versions as technology and software evolve and improve overtime, while preserving previous versions for method reproducibility.
Are there new projects you can talk about?
It’s no surprise that during the pandemic, the top-viewed protocol in the Coronavirus Method Development community is a protocol for sequencing the SARS-CoV-2 genome. It has nearly 175K views and has been forked almost 70 times (i.e., copied and modified by other users). This user engagement really shows the power of an open access platform for sharing public health-related protocols during a pandemic.
FDA’s GenomeTrakr team is currently developing a new wastewater surveillance program for monitoring SARS-CoV-2 variants across the US. This pandemic response project requires rapid development of new methods and associated protocols, which in turn must be shared quickly with our network of collaborating laboratories.
Because wastewater surveillance for viruses is such a new space for public health, we foresee there being broad interest in testing and providing feedback on these protocols, both from public health partners and from academia. The Protocols.io platform enables us to immediately share these pandemic response protocols with the large “Coronavirus Method Development Community” workspace, alerting that broad community of our new protocols.
We, at FDA GenomeTrakr, have found great value using this platform for hosting our protocols. It’s in alignment with our support for open genomic epidemiology data and analysis tools.
I hope to see the protocols.io platform evolve and integrate more with scientific journals, enabling scientists to publish protocols and methods on a platform where these materials can evolve over time while still staying relevant to communities that rely on them.
It’s a great time to be doing science! Let’s make the most of these shared resources.
Take a look at the full set of open access Springer Nature protocol chapters that have been added to protocols.io here and explore the dynamic functionality offered by the platform.
Ruth E. Timme, Ph.D. is a Research Microbiologist at the Office of Regulatory Science at the Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration.
This post was originally shared on The Source.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in