Seeing the individual diversity of filament structures in the forest of supramolecular amyloid assemblies
Published in Chemistry

Amyloid fibrils are ordered nano-structures assembled from proteins or peptides. They are typically less than 10 nm wide but can be up to several micrometres long. They are of interest as novel biomaterials because they can be as strong as steel and as flexible as silk. They are also of great medical interest because some amyloid fibrils are associated with neurodegenerative diseases such as Alzheimer’s and Parkinson’s diseases. The assembly of amyloid, even from a single type of precursor, can result in highly heterogeneous molecular mixtures, as well as a spectrum of different individual filament structures called polymorphs. We believe that individual amyloid particles can display their own biological, physicochemical and materials properties that differ significantly to those of the population average, and this may be related to the varying types of biological response they elicit. It is even possible that individual amyloid particles in molecular mixtures differ in their ‘fitness’, resulting in an environment-dependent selective pressure for the retention of specific polymorphic forms. This could in turn result in the development of resistance to environmental changes such as changes in the solution conditions of their assembly, fluctuations in the biological environment they reside in, or to therapeutic molecules designed to combat their presence in disease.
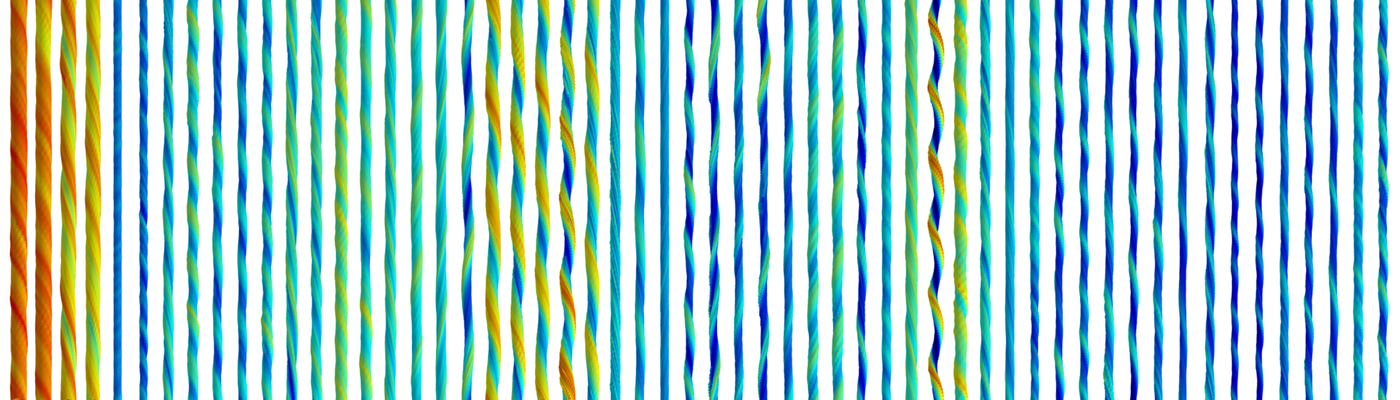
The precise nature, roles and extent of amyloid polymorphism is not yet known. Recent advances in cryo-electron microscopy (cryo-EM) have resulted in the recognition that amyloid polymorphism is indeed omnipresent in human diseases, with the distinct pathology of an amyloid disease linked to a very specific amyloid polymorph or polymorphs. These advances inspired us to search for a way to image, reconstruct, and classify the three-dimensional (3D) supramolecular structures of individual amyloid filaments. In the work communicated in Aubrey et al.: “Quantification of amyloid fibril polymorphism by nano-morphometry reveals the individuality of filament assembly”1 (https://rdcu.be/b64Ar), we applied our recent methodological developments in atomic force microscopy (AFM) image analysis2 and demonstrated that 3D reconstruction of the surface envelope of individual filament structures in a heterogeneous mixture, based directly on the 3D-coordinates obtained from images collected using gentle AFM imaging, is possible. We also devised a multivariate approach to quantify and classify the structural variation of amyloid fibrils within a population, enabled by the 3D structural reconstruction of individual filaments. These advances show directly that the extent of amyloid polymorphism is amino-acid sequence-dependent, and that each and every amyloid particle is a distinct individual member of the ‘amyloid forest’.

Our report also highlights AFM as a superb but often overlooked tool for structural biology. It is over 30 years since Gerd Binnig and Heinrich Rohrer shared half of the Nobel prize in physics "for their design of the scanning tunnelling microscope" (i.e. the predecessor of AFM) with Ernst Ruska "for his fundamental work in electron optics, and for the design of the first electron microscope". Cryo-EM has revolutionised molecular structural analysis since then (cf. the Nobel prize in Chemistry in 2017). Yet, despite giant improvements in commercial and very accessible AFM hardware, AFM has remained a niche method for structural biology. This is due to lack in the development of how the 3D information from AFM topographic image data are understood, extracted and used. Our work1,2 has shown that one can apply AFM to structural analysis beyond mere nanoscale imaging, and brought in its unique advantages of relatively simple physics and the ability to capture individual molecular variations with its phenomenal signal to noise ratio. We fully expect these ideas to lead to future developments that bring together AFM, cryo-EM and other methods in integrated structural biology methodologies that will allow the visualisation of not just the average structure to atomic resolution, but also individual molecules in the ‘amyloid forest’ and how they individually evolve with time.
1. Aubrey, L. D., Blakeman, B. J. F., Lutter, L., Serpell, C. J., Tuite, M. F., Serpell, L. & Xue, W.-F. Quantification of amyloid fibril polymorphism by nano-morphometry reveals the individuality of filament assembly. Commun Chem 3, 125 (2020). https://doi.org/10.1038/s42004-020-00372-3
2. Lutter, L., Serpell, C. J., Tuite, M. F., Serpell, L. & Xue, W.-F. Three-dimensional reconstruction of individual helical nano-filament structures from atomic force microscopy topographs. Biomol. Concepts 11, 102-115 (2020), https://doi.org/10.1515/bmc-2020-0009
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Advances in Asymmetric Catalysis for Organic Chemistry
Publishing Model: Open Access
Deadline: Mar 31, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in