To study a primordial nervous system, Leonid Moroz brings the tools of biology to the open sea. Nature Methods spoke with the neurobiologist turned sea adventurer.
-Ship-Seq is a boat with a sequencing lab on board. On the high seas, Moroz and his crew of sailor-scientists do high-throughput sequencing of DNA and RNA from single cells, as well as neurobiology experiments. And they analyze results, too.
He is especially intrigued by ctenophores, now believed to be the first multicellular organisms, which also have a nervous system but it is utterly unlike ours. It is likely, he says, that their ‘elementary brains,’ their neural and muscular systems, such as the ones found in molluscs and basal metazonas, have evolved independently from all other animal lineages.
In his Nature paper recently published, he and his colleagues present the genome of the ctenophore of the Pacific sea gooseberry (Pleurobrachia bachei)—the data are here—along with transcriptome analysis of other ctenophores. He and his colleagues also present metabolic and physiological data about these organisms. The authors describe how ctenophores have evolved neuronal organizations that show ‘molecular innovations.’ There is also an accompanying News and Views piece by Andreas Hejnol of the University of Bergen in Norway and a Nature news story by Ewen Callaway.
Although organisms can be taken from the sea to the lab, they often need ocean depths or a certain temperature to survive. And when samples are prepared for travel, they need optimized conditions to not degrade. Three decades of dealing with dead organisms, degraded samples, delayed shipments and customs snafus have led Moroz to try something new: Ship-Seq. “We cannot bring the sea to the lab, but we can bring a whole lab to the sea,” he says.After completing two proof-of-concept Ship-Seq voyages—one to the Bahamas and another near the Florida Keys and one to Palau to prepare those voyages—Moroz shares some of his findings here, offers a glimpse at his logistics and future plans. He hopes others can follow his example, because probing and analyzing nature while in and around nature is an adventure with biomedical value.
Biologist and entrepreneur Craig Venter and his Global Ocean Sampling Expedition in some ways parted the seas for Moroz’s project. Moroz wanted to explore biodiversity through sequencing but also take an extra step to do on-site ‘integrative experimental biology,’ which is about using many types of tools to study whole organisms, their behavior and their cells and genomes.
Field biology tends to be an observational science, because in the field, biologists do not usually have an entire high-tech molecular biology lab in tow. And, says Moroz, field scientists may not be completely familiar with new genomics tools, which is too bad since nature has performed genetics experiments waiting to be evaluated. On the boat he studied regeneration, which is hard or even impossible to accomplish “in a dish,” he says, because the animals he studies are incredibly fragile.
King of Regeneration
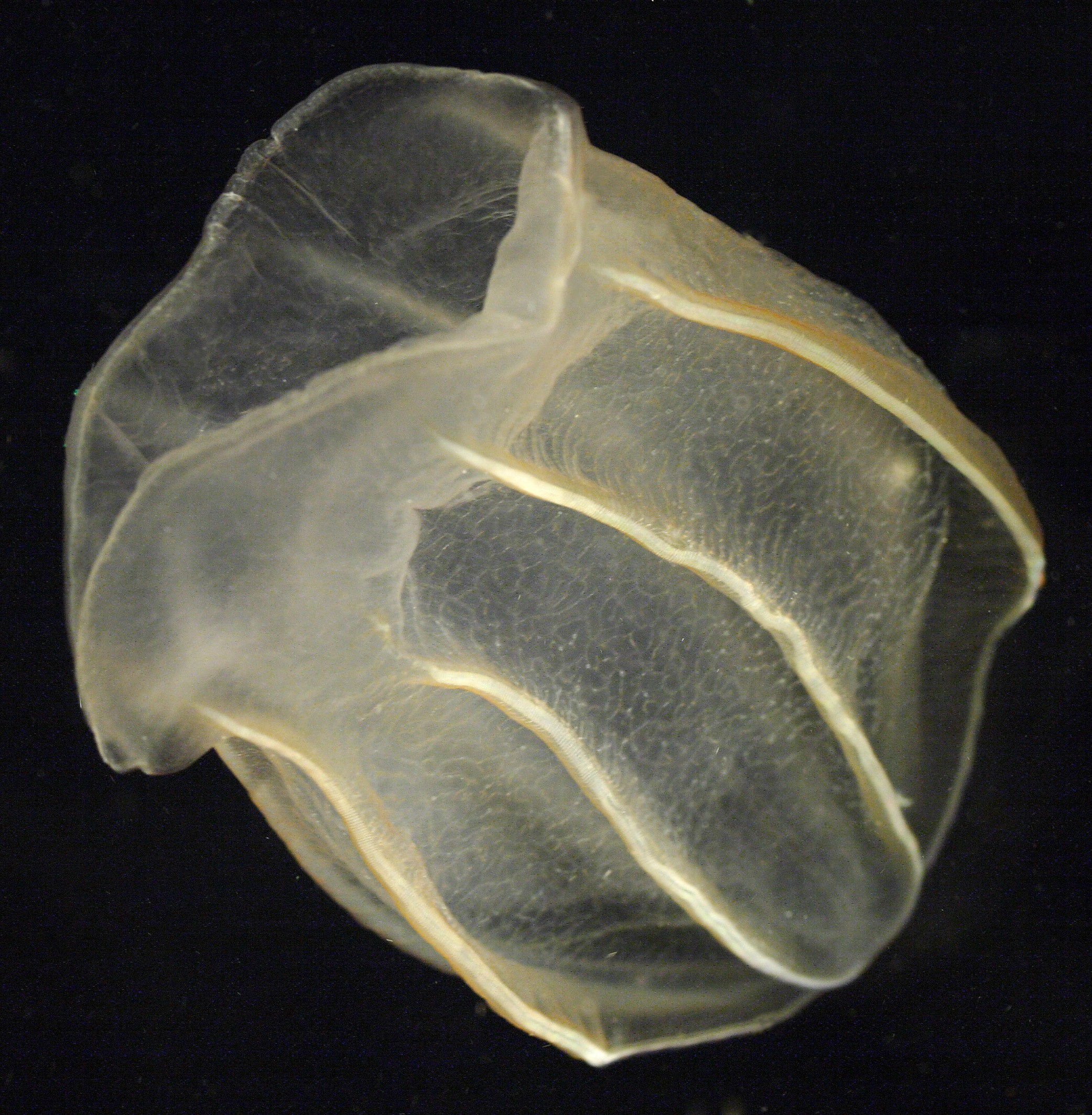
Meet the comb jelly Bolinopsis, which Moroz calls ‘the king of regeneration.’
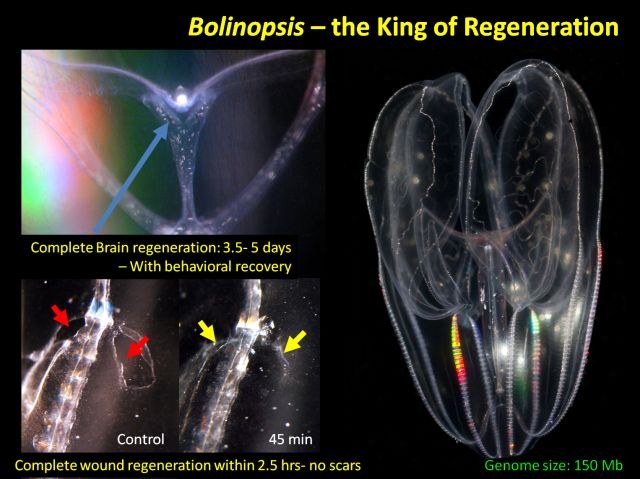
These transparent organisms from the phylum Ctenophora propel themselves through the water with rows of iridescent combs of tiny hairs. Though they may be small and unassuming, they perform an amazing feat: they can regenerate their entire ‘elementary’ brain in three to five days.
Moroz calls their aboral organ with gravity sensors an ‘elementary’ brain; it is not homologous to the human brain. But it is a control center with many neuron types and it coordinates behaviors and motions. In that sense it is an “analog” of the human brain, he says. What astounded Moroz is that when it is dissected from the animal, it grows back.
Other marine organisms such as Hydra are known to regenerate organs, but examples are limited, particularly for organisms that can be maintained in the lab. Finding models for such biological phenomena are crucial in neurobiology, he says. And for regenerative medicine, too. Aplysia, the marine sea slug, has long been helping scientists study memory. And there are more such organisms to find and with which he wants to do ‘real-time’ experiments and analysis, for example look at the dialogue between pre- and post-synaptic neurons.
Bolinopsis has another intriguing trait that Moroz discovered by accident. He was making some small incisions and then briefly interrupted his work. “When I came back around 40 minutes or an hour later, I couldn’t find my cut,“ he says. He made another incision and watched the wound begin to close before his eyes. Overnight, the wound became invisible. “It’s very cool,” says Moroz.
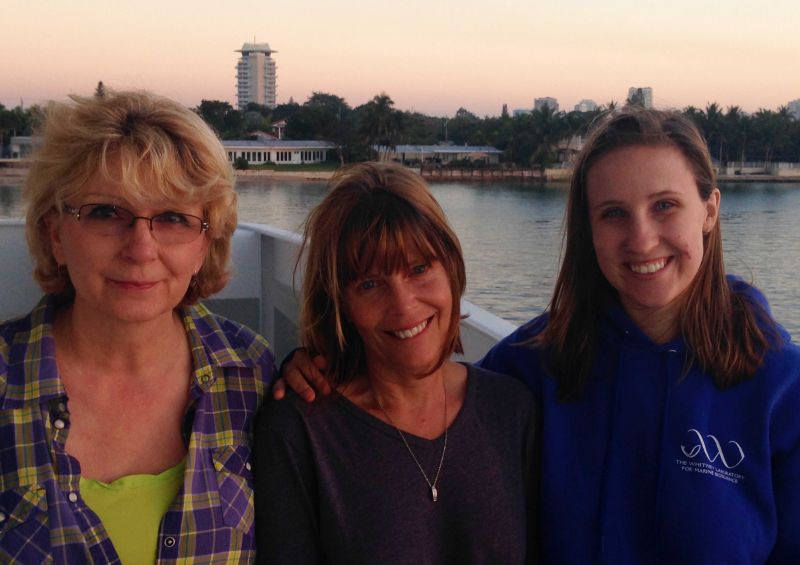
Sequencing team on the first ShipSeq voyage, from left to right: Tatiana Moroz, Andrea Kohn, Rachel Sanford (L.L. Moroz)
Traditional ways of exploring the biochemical underpinnings of physiology and behavior can be slow. With new technologies such as high-throughput sequencing, it is possible to connect data types more quickly. For example, one can see an organism behave and use genomics to see molecular changes, for example in gene expression or epigenetic markers. Being on the boat lets scientists directly address observed biology; “you basically follow up with what nature suggests to you,” says Moroz.
One-way ticket

The Ship-Seq sequencing team for the second trip (from left to right Suzette,Lauran, Rachel, Gabby, Andrea, Greg, Emily, Leonid, Gustav). (Photo: L.L. Moroz)
Comparative biologists face the criticism that their work does not have ‘translational value’ for biomedicine. But Moroz believes Ship-Seq shows that marine organisms have tremendous biomedical value. Bolinopsis is one example of many.

A small volcanic island in Antarctica. Moroz nicknamed it Aplysia Island because it looks like the sea slug, Aplysia, a model organism. (L.L. Moroz)
By delivering the basic alphabet of an organism, sequencing is a boon to many fields. What scientists also need is the grammar with which this alphabet creates the biological equivalent of language, which is behavior and physiology.
With his approach to ‘real-time genomics,’ he wants to help expose this grammar, says Moroz. For example, scientists might want to capture epigenetic changes over the course of learning or regeneration.
Ship-Seq logistics
-
This is Leonid Moroz’s boat, the Copasetic, a 141-foot yacht. Actually it isn’t his boat. And the story about how he gained access to it, is a tale of Moroz’s brand of determination.
Logistics expenses for field expeditions are usually not covered by traditional grants, so Moroz built a collaboration between companies and non-profits to make Ship-Seq a reality.Over the years, he found opportunities, but the tide was against him. One time, everything was ready to go, but the boat’s owner decided to sell the boat, a mere week before the scientists wanted to set sail. Ship-Seq’s maiden voyage was cancelled.
Then Moroz came across the Florida-based International Seakeepers Society, through which yacht-owners loan out their boats for research purposes when they are not using them.
In late 2012, Moroz was invited to an International Seakeepers Society dinner. He had a semiconductor chip in his pocket that is used in semiconductor-based sequencers from Life Technologies, now a part of Thermo Fisher. The scheduled presentation was delayed due to a glitch with the projector. Until the projector was fixed, Moroz gave an impromptu talk about how the small chip could help save the oceans’ heritage and tell the world about the genomic blueprints of marine organisms. He had already been using the technology in his lab and saw how the instrument was accelerating his work.
Some of the listeners smiled politely and ignored him, he says, but a few were excited. Around nine months after that dinner, finally an opportunity presented itself that allowed Ship-Seq to leave the dock.
Boat, crew, captain
Steven Sablotsky, a University of Florida alumnus, engineer, businessperson, yacht owner and member of the International Seakeepers Society approached Moroz. Sablotsky had designed his own boat, the 141-foot Copasetic, with marine research in mind. Sablonsky offered his boat for Moroz’s “proof-of concept” trips for free, including his crew.The added crew was important. Private boat owners can be their own skippers, but large boats are legally obliged to have a competent crew. “It’s pretty complicated machinery,” says Moroz. “You really have to work around the clock.”
At the time, Moroz was also speaking with sequencer manufacturers. He had set up a Life Technologies’ Personal Genome Machine (PGM), which is a bench-top, semiconductor-based sequencer. The instrument’s semiconductor chip uses millions of wells to capture DNA sequence information. DNA is fragmented and each fragment is attached to a bead, and copied such that each bead is covered with copies of the same fragment. One bead is deposited into each one of many wells on the chip, which is then flooded with one of the four DNA bases. When a base is incorporated into DNA, a hydrogen ion is released, leading to a chemical change in the well. The instrument detects the change, converts the signal to voltage, which registers that the base was incorporated and adds it to the growing sequence of the fragment. Another base floods the wells and the process repeats.
After testing the PGM, Moroz decided that it should be the sequencer for Ship-Seq. He was not sure where to install it along with the other necessary lab equipment. It was the Copasetic’s captain Ian van der Watt who suggested housing the lab in a shipping container. A construction manager at Florida Biodiversity Institute helped to organize one such container and design the mobile lab with Moroz. A few weeks later it was ready to be placed on the boat’s deck.
The advantage of a container, says Moroz, is that it offers a completely controlled environment. He and his lab collected the supplies and instruments they needed such as benches, anti-vibration tables, PCR machine, and enrichment systems to measure RNA and DNA and run quality controls.
They needed a high-quality water purification system for the sequencing. It is, he says “somewhat ironic” that the team needed to produce ‘clean pure water’ even though they were in the middle of the ocean. Thermo Fisher engineers got the sequencer ship-shape for a seafaring environment. “Basically we made a full-scale molecular lab” for genomics and imaging, says Moroz.
He still had concerns about variables such as temperature and vibration. They set up the lab and tested all the instruments. While at the dock, he asked the captain to power the motor forwards and backwards, simulating high waves. The lab aced the test.
ShipSeq set sail on its first voyage and the lab was humming from the moment they left, Moroz says. Sablotsky came along, too. Every day they did two sequencing runs and sent the data via a satellite link to HiPerGator, which is a high performance computer with 24,000 core processing units installed at the University of Florida.
Moroz had set up an analysis pipeline with computational tools and scripts to assemble and annotate the incoming sequence information. After automated analysis, data was beamed back to the boat. The sailor-scientists had considered taking a Thermo Fisher engineer along but that did not pan out “so we were on our own,” says Moroz. The good news was “everything worked.”
The second trip, to the Gulf Stream and Florida Keys was windy and through rough seas. Seasickness immobilized half of the lab staff for part of the trip, says Moroz, including his wife. “People could not cope with the field conditions but the PGM machine could,” he says of the sequencer on board. Actually, he says, the Ship-Seq’s sequencing runs were higher quality than in the lab on land. He speculates that the waves enhanced the mixing of chemicals.
“The versatility of our bench top sequencers is only limited by the imagination of today’s scientists,” says Mark Stevenson, executive vice president of Thermo Fisher Scientific in an e-mail to Nature Methods. “Clearly, Dr. Moroz has taken an ingenious idea to a new level and demonstrated that great data can be attained and analyzed in real time – even on a ship that’s rocking on the high seas.”
Seasick but happy
On both trips and despite the seasickness on the second venture, the lab’s team was especially motivated, says Moroz. “It is easy to work a 16-18 hour day when you have the beautiful sea, beautiful creatures around.” People have been important for the overall success of the venture, he says.
Moroz wants to do more trips and expand Ship-Seq’s scientific scope. Using a prototype of the PII chip (which is not yet on the market), he performed single neuron RNA-sequencing in the lab. He projects it might cost around $3 per individual neuronal transcriptome, if one wanted to do a census of neuronal cell types in the brain of a marine organism such as Bolinopsis or others ctenophores, plankton and other, as he calls them, ‘aliens of the sea.’
Ship-Seq and its ‘lab-in-a-container’ offers many opportunities, he says. “The beauty is that it is mobile.” The container could be put on a ship in Florida or it could be sent to Palau or Antarctica and placed on a boat there for not much greater cost. “You can get anywhere,” he says, maybe even set up a “sequencing fleet.”
The planning for the next Ship-Seq trips is underway—but the geographic and scientific directions are not yet finalized. And the finances, too, need to be organized. The trip might focus on more complex marine organisms. For example, cephalopods have complex brains, lending them their nickname ‘primates of the sea.’ Moroz hopes to one day study their neurobiology, integrating field biology, behavior, and genomics. He also wants to be part of the ongoing ‘race to save species,’ to not only study but also “preserve our planet.”
Moroz has encountered plenty of detractors and skeptics. Whenever he is criticized and told he should stick to the traditional way of doing science, his path of taking the lab to the sea feels right. He says it reinforces his sense: “I must do it.” To him, doing science on Ship-Seq feels like “the investigation of a new planet.”
Ship-Seq Protocol
1 x 141-foot boat
1 x generous entrepreneur
1 x ship’s crew
1 x mobile molecular biology lab equipped with lab benches, a sequencer, reagents
1 x manufacturer of a high-throughput sequencer willing to donate an instrument
1 x satellite link to a supercomputer
1 x lab staff and scientist/wife willing to be scientist-sailors
1 x diving equipment
1 x funding National Institutes of Health (NIH), National Science Foundation (NSF), National Aeronautics and Space Administration (NASA)
3 x support from non-profit organizations: Florida Biodiversity Institute, Florida Museum of Natural History, the International Seakeepers Society
1,000 international units of patience
Several remedies for seasickness













Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in