On this 10th anniversary of the first issue of Nature Methods it is appropriate to look back at the relationship between the journal and super-resolution microscopy, one of the technologies we have chosen as one of the top ten methods developments in the ten years since Nature Methods published it first issue.
Super-resolution microscopy first appeared in Nature Methods with the online publication of two papers on August 9, 2006. One demonstrated the first super-resolution microscopy image using a genetically-encoded fluorescent probe (Willig et. al., 2006) and the other was the first publication describing stochastic optical reconstruction microscopy (STORM; Rust et. al., 2006), the class of methods now often referred to as single molecule localization microscopy (SMLM). Initially, the papers were mostly overshadowed by the media storm accompanying the publication one day later of photoactivated localization microscopy (PALM) by Betzig et. al. in Science, but the STORM and PALM papers together were instrumental in driving wider development of the nascent super-resolution microscopy field that had previously been confined to a small number of highly specialist groups.
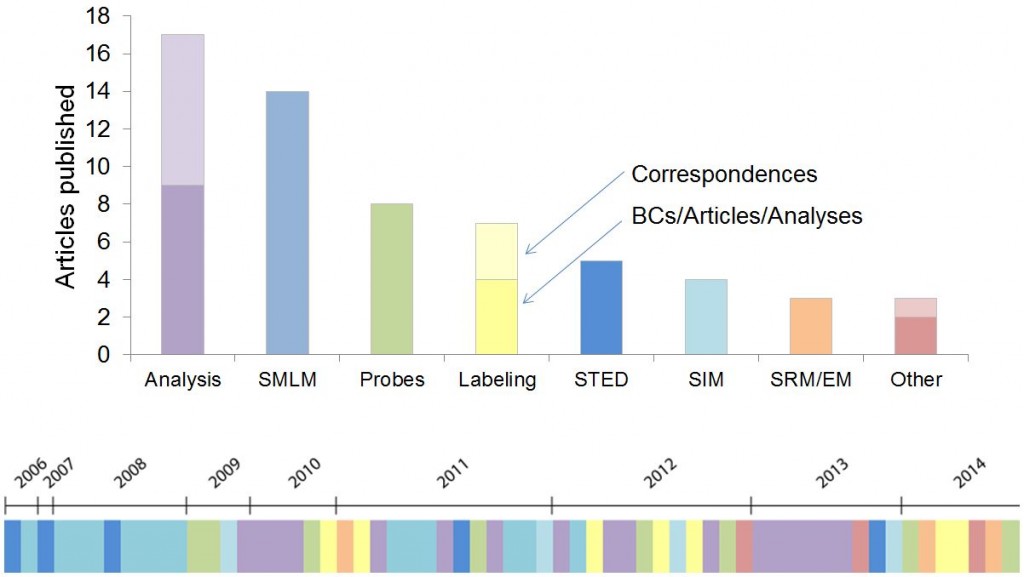
Nature Methods has now published 64 articles on super-resolution microscopy, 49 full original research articles, 11 Correspondences and 4 Review and Commentary articles. The accompanying illustration conveys the wide range of topics covered and their historical progression. Super-resolution microscopy was also our choice of Method of the Year in 2008.
The first three years of super-resolution microscopy (SRM) publications in Nature Methods were dominated by advances in localization-based SRM and early attempts at live cell SRM. Betzig and colleagues defined important considerations for performing live-cell PALM (Shroff et. al., 2008) and PALM was adapted as a massively parallel single particle tracking technique called sptPALM (Manley et. al., 2008). Another early paper demonstrated the use of dual-plane imaging for 3D SRM several microns deep into a sample (Juette et. al., 2008), but to this day SRM is still dominated by 2D imaging.
It was clear that the probes used for localization-based SRM were critical to the performance of these techniques. In early 2009 we published the new fluorescent proteins, PA-mCherry (Subach et. al., 2009) and mEOS2 (McKinney et. al., 2009), from the Verkhusha and Looger labs respectively. The performance characteristics of mEOS2 and the Looger lab’s very open reagent sharing habit helped contribute to this protein dominating much fluorescence protein based SRM.
In late 2009 we began to address the prior lack of sufficient attention to the analysis methods used in localization-based SRM with the publishing of two papers (Mortensen et. al., 2009 and Smith et. al., 2009) focused on minimum likelihood algorithms for precisely estimating the centers of fluorophore image spots, a fundamental underpinning of the whole class of localization-based SRM methods. At this time we also started publishing Correspondences describing user-friendly software for performing the early localization analysis steps; first LivePALM (Hedde et. al., 2009) and QuickPALM (Henriques et. al., 2010) and in later years DAOSTORM (Holden et. al., 2011) and RapidSTORM (Wolter et. al., 2012). During this period researchers also scoured other fields for algorithms new to imaging analysis and imported the powerful compressed sensing analysis method (Zhu et. al., 2012), developed novel localization methods like radial symmetry (Parthasarathy, 2012) and characterized and corrected the noise attributes of sCMOS cameras so that they could challenge EMCCDs as the camera of choice for localization-based SRM (Huang et. al., 2013). A particularly interesting development was the use of Bayesian analysis for image generation that didn’t require explicit fluorophore localization and could work with the intrinsic blinking and bleaching of high density GFP-labeled live samples (Cox et. al., 2011).
We soon determined that the later analysis steps required for interpretation of the underlying biology were most ripe for, and in need of, further development. Improper analysis could easily lead to artifacts, particularly when trying to use localization-based SRM to examine protein clustering (Annibale et. al., 2011). Notable early work in this area was the use of pair correlation analysis to examine protein organization in the plasma membrane (Sengupta et. al., 2011). An ongoing issue in analyzing localization-based SRM images has been determining the resolution of the resulting image, a far less straightforward task than one might expect. Adoption and development of Fourier ring correlation from electron microscopy provided a compelling solution to this challenge (Niewenhuize et. al., 2013) but more work remains to be done before researchers can be confident of reliably measuring the resolution of their images.
Although manuscripts with a focus on analysis methods made up the majority of articles published in Nature Methods over the past 10 years, there were also continuous developments in imaging technology. STED microscopy was improved through the use of continuous wave lasers (Willig et. al., 2007) and time gating (Vicidomini et. al., 2011). A STED configuration that created a spherical scanning spot was used to image the 3D structure of a single mitochondria (Schmidt et. al., 2008). There was also further development of the optical methods used for localization-based SRM. Temporal focusing of two-photon irradiation allowed confined photoactivation in whole cells, thus limiting photobleaching outside the imaging area (York et. al., 2011). Confined photoactivation and imaging was also accomplished using dual orthogonal objectives to combine light-sheet microscopy with localization-based SRM (Cella Zanacchi et. al., 2011). Finally, a dual-objective scheme with objectives facing one another combined with astigmatism improved the resolution of 3D localization-based SRM (Xu et. al., 2012).
In recent years, alternative SRM methods made an appearance. The scanning-based method, reversible saturable optical fluorescence transitions (RESOLFT), was massively parallelized and used for imaging whole living cells (Chmyrov et. al., 2013). An intriguing recent report combined elements of STED and localization-based SRM in a new imaging modality that discriminates nanoareas of fluorescently labeled rigid proteins using polarization (Hafi et. al., 2014).
Improvements in imaging technology are of little use if you can’t label your targets of interest. Labeling methods have therefore been an important component of the SRM papers published in Nature Methods. Trimethoprim labeling (Wombacher et. al., 2010) and SNAP tag labeling (Klein et. al., 2011) both allowed direct labeling of proteins in live cells and this was combined with bright fast-switching probes to allow fast 3D localization-based SRM in whole living cells at ~25 nm resolution (Jones et. al., 2011). Other investigators improved labeling not by direct labeling using chemical tags, but by using smaller affinity probes such as nanobodies (Ries et. al., 2012) or aptamers (Opazo et. al., 2012). A particularly intriguing class of labeling methods relies on DNA oligos. Barcoding (Lubeck et. al., 2012) and sequential labeling (Jungmann et. al., 2014; Lubeck et. al., 2014) allowed highly multiplex labeling of target proteins and nucleic acids.
With so many developments and choices for researchers it is increasingly important for them to have quality data on the relative performance of different techniques and tools. To this end, Nature Methods has been publishing increasing numbers of Analysis articles reporting such performance comparisons and SRM has been no exception to this. The performance of a wide selection of chemical fluorophores for localization-based SRM was characterized in a real-life imaging situation (Dempsey et. al., 2011) and a recent report characterized the photoactivation efficiency of fluorescent proteins (Durisic et. al., 2014).
We hope you enjoyed this brief summary of SRM in Nature Methods. Although I have tried to include much of what Nature Methods has published in this field, the summary is by no means comprehensive. Most significantly, it doesn’t include many of the methods that are used to double the resolution of fluorescence microscopy. If there is sufficient interest we will consider extending our summary to include both these and more recent developments as they occur.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in