The Biogeography of Algal Microbiomes from the Arctic to the Antarctic
Published in Microbiology

Sampling biases shape our view of the natural world and this particularly influences our knowledge of species diversity and biogeography [1]. The latter, though, is instrumental if we want to understand how climate change impacts the distribution of species and communities across space and time. In spite of global efforts to map microbiomes (community of micro-organisms living together in a particular habitat) of the oceans [2], the deep sea and the polar oceans remain poorly studied, impacting our ability to predict biodiversity loss to global changes. As these cold aquatic ecosystems represent the largest habitats on Earth, they can be considered key biodiversity areas, and if we do not include them for comparative analysis with their better studied warm counterparts, we are not only missing a major proportion of global biodiversity but also are likely to misinterpret the role of habitat characteristics in shaping the global distribution, ecology, and evolution of species and their populations [1].
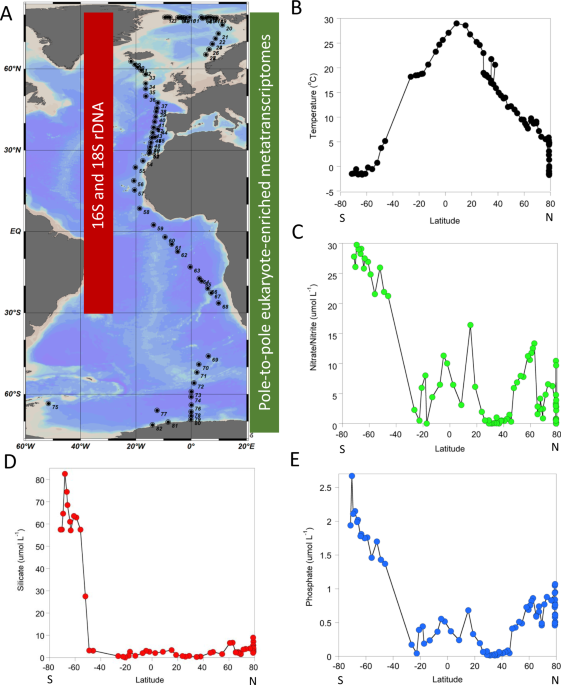
To address sampling bias in terms of estimating the diversity and biogeography of upper ocean phytoplankton microbiomes, in 2012 we began to sequence phylogenetic marker genes (16/18S rDNA) of Arctic phytoplankton communities with a focus on eukaryotic primary producers (microalgae) and their co-occurring bacteria. We complemented this taxonomic work with the sequencing of expressed microalgal genes (metatranscriptomics) to reveal polar-specific metabolic characteristics of oceanic primary producers. This work was mainly enabled by funding from the US Department of Energy Joint Genome Institute (JGI), the Natural Environment Research Council (NERC), and the Alfred-Wegener Institute (AWI) for Polar and Marine Research. RV 'Polarstern', a German icebreaker, was used as a research platform to sample the upper Arctic Ocean until 79°N near the east coast of Greenland. With help of the Polarstern crews, we were able to sample over 100 stations from pole to pole. For most samples, we utilised a CTD Rosette (Fig. 1) sampler allowing us to find the depths with the highest concentrations of phytoplankton.

In the Arctic, we had the opportunity to use a so-called mummy chair (Fig. 2) and get some samples of under-ice communities (Fig. 3) that got swept up as the ice got broken up by Polarstern.

Processing the water samples was challenging in the polar environments as we had to keep them as close to their natural temperature as we could. Thus, filtrations were done in cooled containers to avoid early degradation of their DNA and more importantly their RNA. Over subsequent years, this initial work in the Arctic Ocean was extended to sampling across the Atlantic Ocean and the Southern Ocean with the southernmost samples obtained close to the Antarctic shelf-ice edge at 71°S. In total, four separate cruises between 2011 and 2017 were required to sample algal microbiomes from pole to pole, which built the foundation of our paper. One of them was led by colleagues from the Netherlands who helped to bridge a sampling gap in the North Atlantic. With a large pole-to-pole phytoplankton metatranscriptome sequence dataset and accompanied amplicon (PCR replicated locus of DNA) data sets for microalgae and their co-occurring bacteria, our group was challenged with sequence analyses. Although our sampling and sequencing efforts were largely of exploratory nature, the overarching question of this study was to reveal what drives spatial scaling of changes in the diversity of algal microbiomes, i.e. loss and gain of species but also their expressed genes. This question goes beyond assessing what drives the biodiversity of local microbiomes (local (alpha) diversity) and as far as we can tell has never been addressed before for marine microbiomes and their expressed genes from pole to pole. In order to address our overarching question, we applied two independent analyses: the identification of a) networks to identify who co-occurs with whom across major latitudinal gradients and b) spatial diversity patterns underlying dissimilarities of algal microbiome species compositions between neighbouring samples from pole to pole. The latter concept is also known as beta diversity. Unexpectedly, both analyses independently revealed that there appears to be an ecological boundary in the upper ocean geographically separating polar and non-polar algal microbiome.

The demarcation line was estimated to be at an annual average of 14-15°C in the upper ocean. This temperature separates permanently mixed from permanently stratified upper ocean waters on an annual average and has previously been identified for the geographical partitioning of net primary production of phytoplankton communities [3]. Although the importance of macro (e.g. nitrate) and micronutrients (e.g. iron, zinc) is well known for not only differences in the productivity of algal microbiomes but also their diversity and gene expression, the environmental parameter that best explains their separation into polar and non-polar microbiomes including differences in community gene expression is temperature. Thus, temperature appears to be significantly contributing to how the diversity and gene expression of algal microbiomes is distributed in the global upper ocean with consequences for oceanic food webs and biogeochemical cycles they drive under conditions of global warming. As the separation into the two major algal microbiomes appears to be relatively stable, it suggests that at least some microbes including their interacting partners on either side of the demarcation boundary evolved independently from one another to adapt to the very different environmental conditions. Thus, it remains to be seen if this boundary limits gene flow between microbial populations and therefore contributes to speciation. If confirmed, the structure and dynamic of the physical oceanic environment is significantly contributing to shaping microbial evolution, diversity, and activity across space and time, which has not been addressed much yet [4] but might be important in the context of global warming.
References
- https://onlinelibrary.wiley.com/doi/10.1111/ecog.05926?af=R
- https://www.science.org/lookup/doi/10.1126/science.aac5605
- https://www.nature.com/articles/nature05317
- https://onlinelibrary.wiley.com/doi/10.1111/mec.15554
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in