The untold story of TMOD2 in colorectal cancer metastasis
Published in Biomedical Research
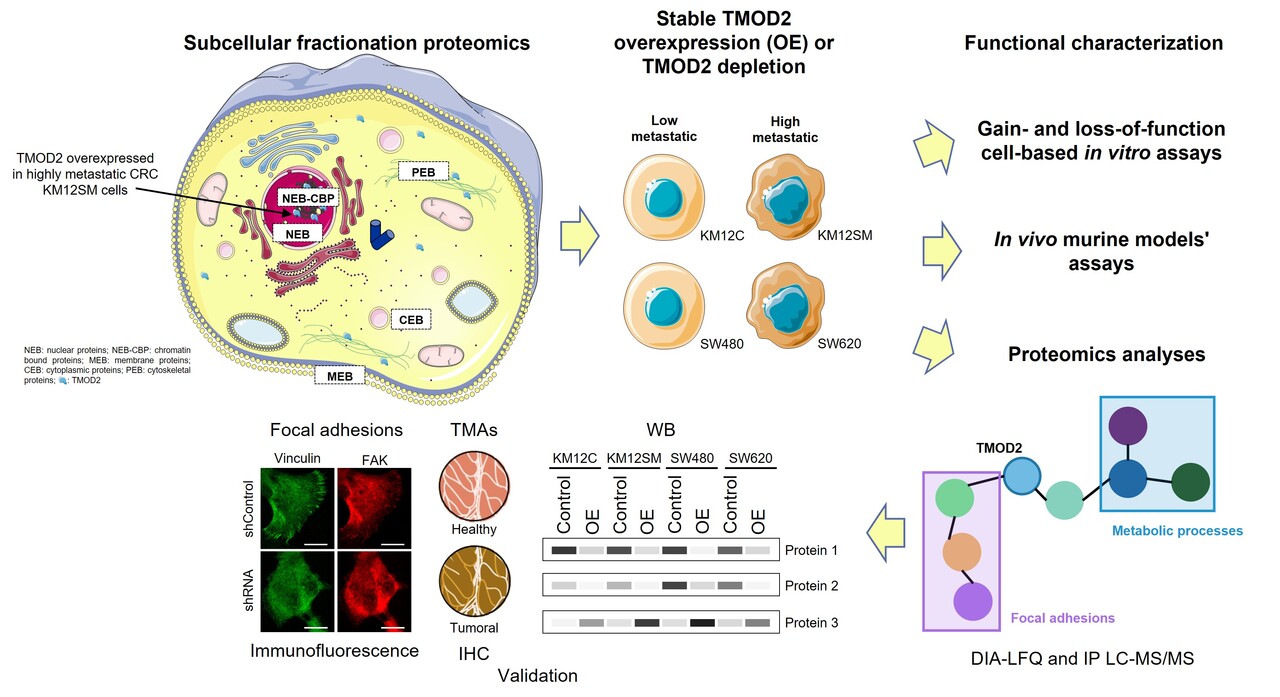
From nuclear proteomes to a hypothesis
TMOD2 had been primarily associated with neuronal function, with only a handful of studies hinting at any role beyond the nervous system. However, TMOD2 emerged as a surprising but interesting and persistent candidate in our search for drivers of metastasis through the twists and turns of proteomics and bioinformatics.
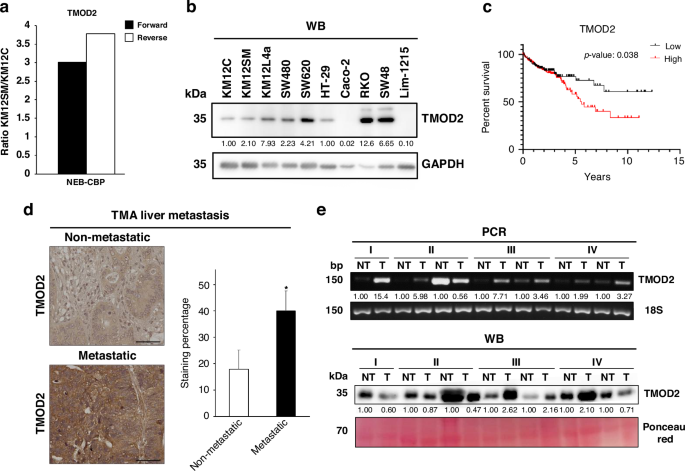
With the objective to identify proteins not previously related to CRC with potential as therapeutic targets or biomarkers of the disease, previous researchers studied the KM12 CRC cell model -a well-established model of CRC metastasis-. This model is composed of the poorly-metastatic KM12C cells and its isogenic highly-metastatic to liver KM12SM cells. By performing spatial proteomics, the authors were able to identify differentially expressed proteins in the different compartments of these cells (10.1002/pmic.201700094). Among them, TMOD2 consistently stood out as highly expressed in the nuclear fraction of KM12SM cells.
But... why would a protein best known for regulating actin filament length in neurons be upregulated in the nuclei of metastatic CRC cells? Could TMOD2 be playing a broader role in cytoskeletal and nuclei dynamics or in cancer biology?
After analyzing colon adenocarcinoma and rectal adenocarcinoma patient datasets from The Cancer Genome Atlas, we found that high TMOD2 expression correlated with advanced CRC stages and poorer overall survival. Additionally, we also found higher TMOD2 mRNA and protein levels in tumoral samples using an independent cohort of paired tumoral and non-tumoral tissue samples from CRC patients. This convergence of experimental and clinical data sparked our central hypothesis: TMOD2 might not just be a bystander, but an active promoter of CRC progression.
Building a multi-layered approach
To address this, we combined different synergic strategies:
- Functional proteomics (label-free quantification data independent acquisition (DIA-LFQ) and immunoprecipitation (IP) coupled to mass spectrometry (LC-MS/MS)) profile the proteome and interactome associated with TMOD2 and identify the protein changes produced by its induced overexpression.
- Bioinformatics to connect TMOD2 expression patterns with clinical outcomes and biological pathways.
- Cellular and molecular assays to determine how TMOD2 influences proliferation, adhesion, migration, invasion, and anchorage-independent growth.
- In vivo models to analyze the ability of TMOD2 to enhance tumor growth and colonization of distant organs.
What did we discover about TMOD2 in CRC?
To characterize TMOD2 role in CRC, we induced both the stable overexpression and the stable depletion of TMOD2 in two isogenic CRC cell models of metastasis. We employed the KM12 and the SW cell systems, with the second model consisting of the poorly metastatic SW480 CRC cells and the lymph node–metastatic SW620 CRC cells.
When TMOD2 was overexpressed, we observed that CRC cells became more adhesive, migratory, invasive, and capable of inducing anchorage-independent growth, consistent with the acquisition of a more aggressive phenotype. Notably, , these changes were more pronounced in the poorly-metastatic KM12C and SW480 CRC cells, in line with their lower basal levels of TMOD2 compared with their highly-metastatic counterparts. Conversely, TMOD2 stable depletion induced the opposite effects in the tumorigenic and metastatic cell properties.
Additionally, in vivo experiments strengthened and validated these findings. Nude mice inoculated in the flank with TMOD2-overexpressing cells developed larger subcutaneous tumors. More importantly, intrasplenic inoculation of low-metastatic KM12C cells overexpressing TMOD2 led to liver colonization and tumor progression, confirming a metastasis-driving role for TMOD2 in CRC.
How does proteomics support these results?
If TMOD2 was acting as a central player of CRC progression, what were its partners in crime? To address this, we come back to proteomics using the CRC cell lines stably overexpressing TMOD2. Using DIA-LFQ and IP-LC-MS/MS, we mapped proteins dysregulated by TMOD2 overexpression and its potential interactors.
Proteomics results highlighted cytoskeletal regulators, secretory proteins, and focal adhesion molecules. This was consistent with the behavioral changes observed in vitro and in vivo with CRC cells. Taking into account our previous results and the described role of TMOD2 as a regulator of the actin cytoskeleton, we sought to further corroborate the role of TMOD2 in focal adhesions, a key feature in regulating cell adhesion, migration, and metastatic potential. By immunofluorescence, we observed how TMOD2 overexpression contributed to their formation in the poorly metastatic KM12C cells, whereas its stable depletion significantly reduced the capacity of highly-metastatic KM12SM cells to form focal adhesions.
Altogether, these results allowed us to connect the dots between TMOD2’s presence in the nuclear proteome and its impact on cellular architecture. What began as an odd surprising nuclear TMOD2 enrichment ended up revealing a broader role for TMOD2 as a coordinator of metastasis-relevant networks.
The challenges behind the data
Behind the results, there were many challenges. Working with isogenic CRC lines requires meticulous care to avoid confounding factors. Proteomics experiments demand not only technical precision but also careful bioinformatics interpretation; and in vivo metastasis models, while powerful, require long timelines, close monitoring, and ethical responsibility.
But… why TMOD2 matters?
Our findings suggest that TMOD2 could be a promising target for intervention. By modulating cytoskeletal dynamics, enhancing cell adhesion, and promoting liver metastasis, TMOD2 occupies a key position in the metastatic cascade. Inhibiting TMOD2, or its downstream partners identified in this work, could blunt the metastatic potential of CRC cells.
Moreover, the correlation between TMOD2 expression and patient survival indicates that it could serve as a prognostic biomarker. This dual role — as a marker and as a potential target of therapeutic intervention— makes TMOD2 particularly compelling.
However, this story is not finished as the work raises more questions:
- Can safer and more effective in vivo TMOD2 inhibitors be developed?
- Is TMOD2’s role unique to CRC, or does it extend to other cancer malignancies?
These questions will guide the next steps in our research.
Follow the Topic
-
British Journal of Cancer

This journal is devoted to publishing cutting edge discovery, translational and clinical cancer research across the broad spectrum of oncology.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in