It is now possible to map the activity of nearly all the neurons in a vertebrate brain at cellular resolution in just over a second. What does this mean for neuroscience research and projects like the Brain Activity Map proposal?
In an Article that just went live in Nature Methods, Misha Ahrens and Philipp Keller from HHMI’s Janelia Farm Research Campus used high-speed light sheet microscopy to image the activity of 80% of the neurons in the brain of a fish larva at speeds of a whole brain every 1.3 seconds. This represents—to our knowledge—the first technology that achieves whole brain imaging of a vertebrate brain at cellular resolution with speeds that approximate neural activity patterns and behavior.
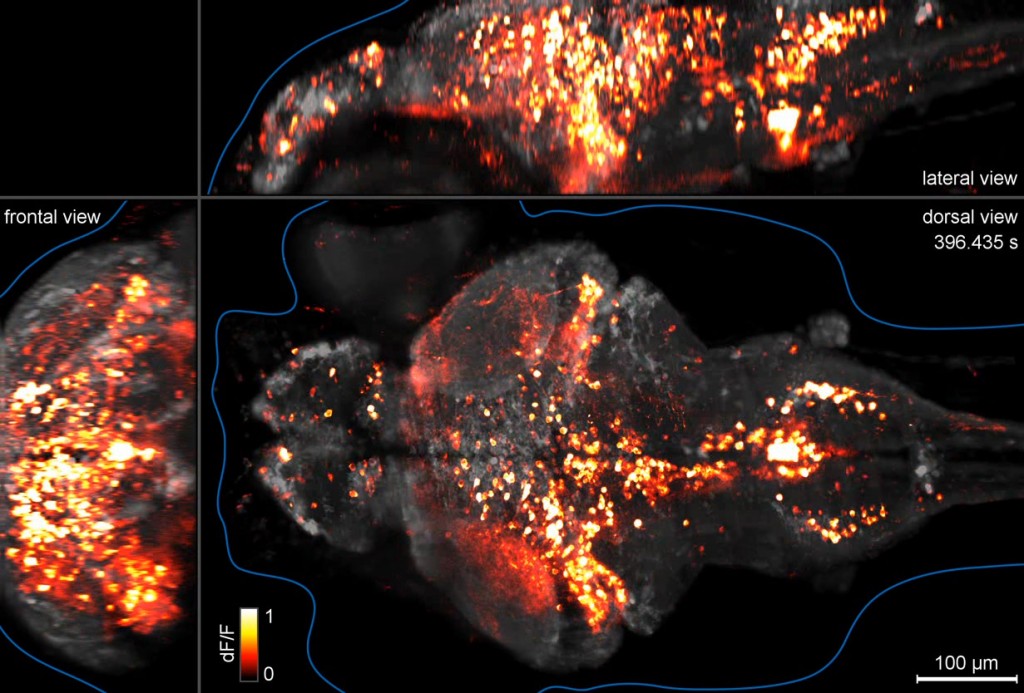
Brain activity imaging of a whole zebrafish brain at single-cell resolution. Click on image to view video [20 MB].
The details of BAM’s exact goals and a clear roadmap and timeline to achieve them have yet to be presented, but from what its proponents have described in a recent Science paper the main aspiration of the project is to improve our understanding of how whole neuronal circuits work at the cellular level. The project seeks to monitor the activity of whole circuits as well as manipulate them to study their functional role. To reach these goals, first and foremost one must have technology capable of measuring the activity of individual neurons throughout the entire brain in a way that can discriminate individual circuits. The most obvious way to do this is by imaging the activity as it is occurring.
With improvements in the speed and resolution of existing microscopy setups and in the probes for monitoring activity, exhaustive imaging of neuronal function across a small transparent organism was bound to be possible—as this study has now shown.
The study has also made interesting discoveries. The authors saw correlated activity patterns measured at the cellular level that spanned large areas of the brain—pointing to the existence of broadly distributed functional circuits. The next steps will be to determine the causal role that these circuits play in behavior—something that will require improvements in the methods for 3D optogenetics. Obtaining the detailed anatomical map of these circuits will also be key to understand the brain’s organization at its deepest level.
These are some of the types of experiments described in the BAM proposal and they are clearly within reach in the next 10 years–whether through a centralized initiative or through normal lab competition and peer review. While it is expected that in mice, too, functional circuits will span large brain areas, performing these types of experiments in mice will require more methodological imagination. It will not be possible to place a living mouse brain within the microscope system used by Ahrens and Keller to image the zebrafish brain. The mouse brain is significantly bigger, is largely impenetrable to visible light and is surrounded by a skull. Realistically, we may not see methods that enable whole brain activity mapping in mammals at the cellular level for quite a while.
But there is much worth learning about brain function in smaller organisms such as the zebrafish and drosophila, and microscopy systems such as this will be capable of providing important fundamental insights into brain function that are relevant to our understanding of the human brain.
Whether it will be through BAM or not, the neuroscience community has important challenges to tackle ahead. At Nature Methods, we have been actively involved in supporting technology development in the neurosciences from the very beginning and we look forward with enthusiasm to doing so during this exciting period in neuroscience research.
Update: We just published an Editorial on this topic in our May issue.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in