A collaborative effort reveals deadly malaria parasites’ strategy for priming red blood cells for invasion
Red blood cells (RBCs) are the body's lifeline, but they also serve as the perfect hosts for one of the deadliest infectious organisms: the malaria parasite Plasmodium falciparum. Malaria is a life-threatening vector-borne disease that ultimately kills about a thousand children around the world every day. Our findings uncovered what enables the malaria parasite to mount such an effective takeover.
The results presented in this paper are the outcome of the merger of the research fields of proteases, extracellular vesicles (EVs) and malaria, showcasing how personal relationships are a powerful force for scientific discovery. The thread that led to this paper can be traced back to when we, Prof. Michal Sharon and Dr. Neta Regev-Rudzki, first met, which was when Michal, an expert on proteasomes and mass spectrometry (MS), mentored Neta, then a new PI who was establishing a lab on the biological aspects of malaria with a focus on EVs. We became good friends, but our research directions remained far apart. That is, until we discussed Neta’s recent discovery that EVs released by the malaria parasite Plasmodium falciparum while in human red blood cells contain, among other factors, proteins of the proteasome.
It is well known that pathogens alter the host membrane to mediate their invasion by activating communication mechanisms that target the host cell. Yet, it was not known whether malaria parasites, while growing inside RBCs, remodel naïve surrounding RBCs in preparation for invasion. Given Neta’s findings, we speculated that P. falciparum uses the EVs to shuttle assembled and active proteasome complexes to neighboring RBCs, where they remodel the naïve RBCs’ membrane so as to ease the subsequent parasites’ penetration.
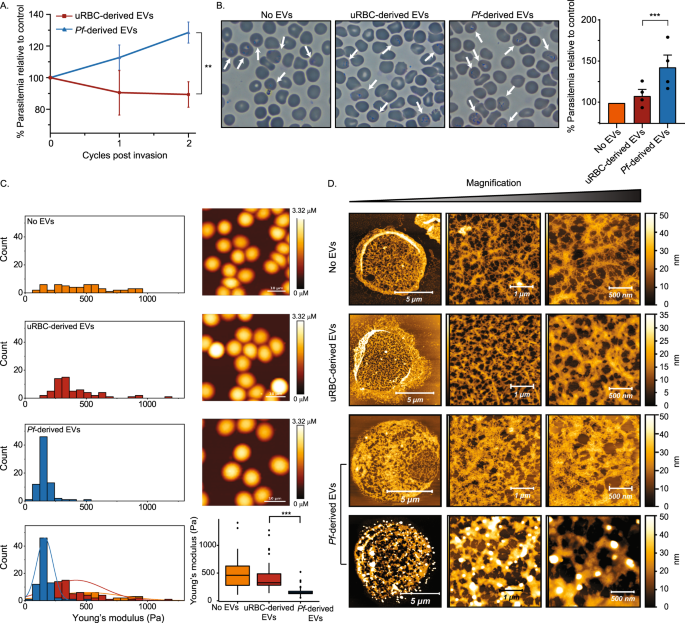
By joining forces, we managed to expose the details of this pre-invasion strategy. First, we established that P. falciparum EVs contain a specific proteasome variant known as 20S, which uses less energy and is smaller than other versions. Working with Dr. Irit Rosenhek-Goldian and Dr. Sidney R. Cohen, we developed a special visualization technique based on atomic force microscopy, which showed that the 20S proteasome pokes holes in the RBCs' cytoskeleton and dissolves some of its filaments. We also identified four RBC proteins targeted for degradation by the proteasome. We further developed a machine-learning algorithm that distinguishes the cytoskeletons of red blood cells that had been altered by the secreted 20S from those that hadn't.
To confirm that the remodeling of the RBCs was indeed due to the activity of the parasitic proteasomes, we blocked the active site of the 20S proteasome with an inhibitor prior to exposing RBC cells to the P. falciparum EVs. This inhibition prevented the remodeling of the cellular membranes, limiting the invasion of the cells by the parasite and, consequently, the parasite's growth.
This last result opens up a new direction in the search for malaria therapies: blocking the parasite's proteasomes so as to reduce its invasion into red blood cells. Our findings may also prove relevant to other infectious diseases, because subunits of the 20S proteasome have been found in extracellular vesicles released by different classes of parasites, for example, those causing sleeping sickness and leishmaniasis. Moreover, the results may be applicable to cancer, as the 20S has recently been detected in extracellular vesicles released by human cells that facilitate tumor growth.
Another takeaway from this study is that, just like “it takes a village to raise a child,” our study could not have progressed without the collaboration of researchers from many different fields—not only our respective specializations, but also bio-physicists, computational biologists, imaging, microscopy and proteomic specialists. The research team included Dr. Elya Dekel, Dr. Dana Yaffe, Dr. Gili Ben-Nissan, Dr. Yifat Ofir-Birin, Dr. Mattia Morandi, Yael Ohana Daniel, Paula Abou Karam, Daniel Alfandari, Shimrit Malihi, Tal Temin Block, Dr. Or-Yam Revach, Ariel Rudik and Dr. Ori Avinoam of Biomolecular Sciences Department; ; Dr. Ron Rotkopf, Dr. Ido Azuri and Dr. Ziv Porat of the Life Sciences Core Facilities Department; Debakshi Mullick and Prof. Nir S. Gov of the Chemical and Biological Physics Department; Dr. Tamar Ziv of the Technion – Israel Institute of Technology; Dr. Xavier Sisquella, Dr. Matthew A. Pimentel, Dr. Thomas Nebl and Dr. Eugene Kapp of the University of Melbourne and Walter and Eliza Hall Institute of Medical Research; Giulia Bergamaschi and Prof. Gijs J.L Wuite of Vrije Universiteit Amsterdam; Dr. Raya Sorkin of Tel Aviv University; and Dr. Teresa Carvalho of La Trobe University, Melbourne.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in