Phenotypes and progression pathways in severe malaria
Published in Healthcare & Nursing
Severe malaria causes over 400 000 deaths a year, mainly in African children. Malaria is a complex disease, with different symptoms appearing at different times in different patients. This makes it hard to assign risks and optimise treatments for individual patients. The current World Health Organisation (WHO) guidelines take a very “broad-strokes” approach, based on a small number of key features. Unfortunately, treatments based on these guidelines may experience limited success – malaria can vary substantially patient-to-patient, and what works for one may not work for others.
In this project, we asked whether we could use data science and machine learning to gain more detailed and predictive insight about malaria progression and prognosis. We work in a “precision healthcare” paradigm, using mathematical approaches to harness a large dataset on clinical malaria presentations. This dataset included incomplete observations of the clinical presentations of 2 904 children with severe malaria, as well as their survival outcome. 38 features were recorded, of many different types, ranging from simple “is the patient coughing?” to detailed measurements of haemoglobin and parasite levels in blood.
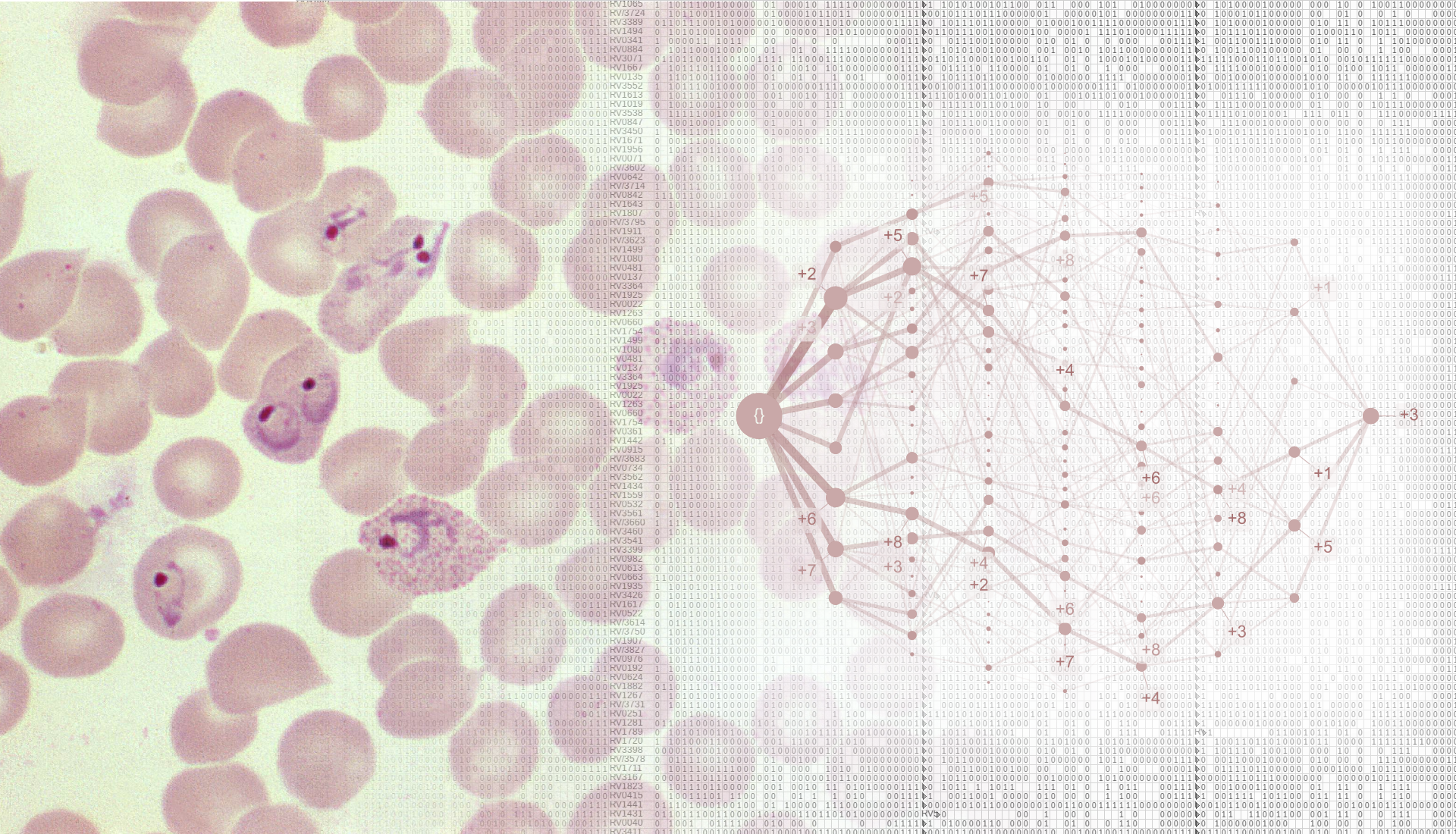
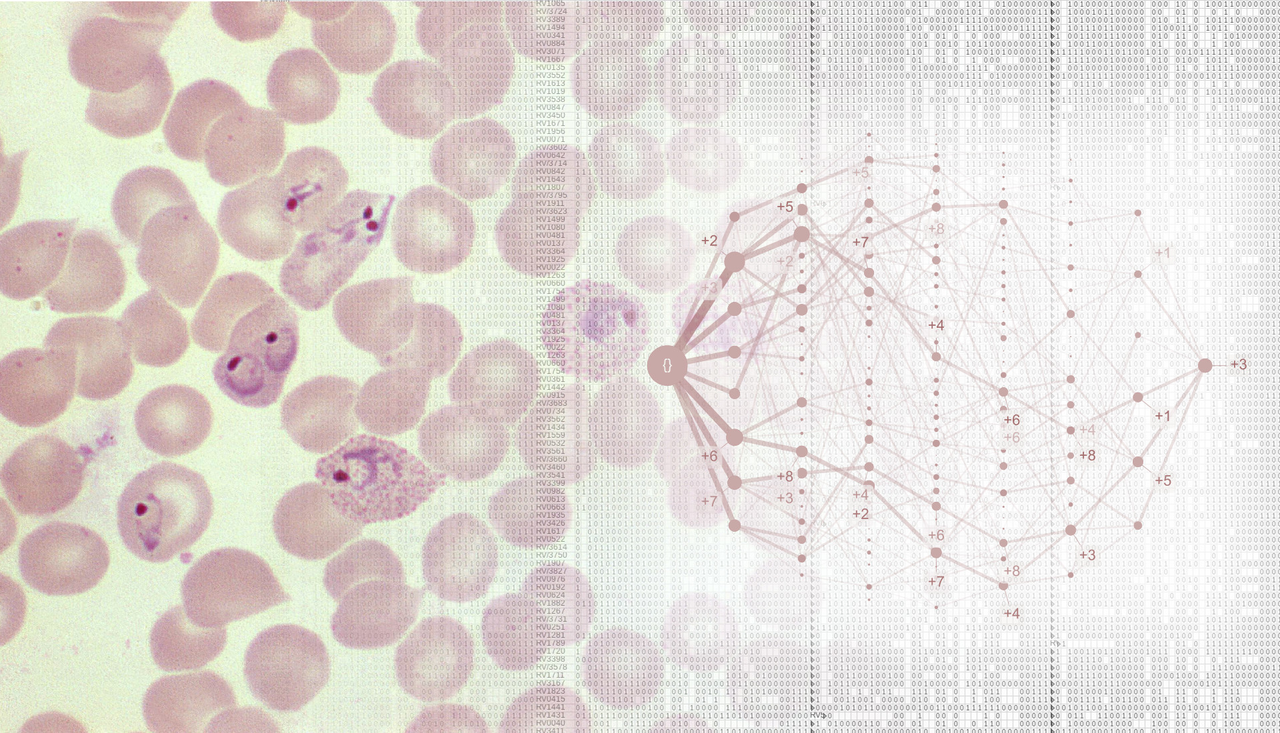
Severe malaria is a devastating disease caused by Plasmodium parasites (left). We used a large-scale dataset on malaria symptoms in individual patients (right, background) together with cutting-edge algorithms (including “HyperTraPS”, right, foreground) to identify high-risk symptom sets and malarial progression pathways.
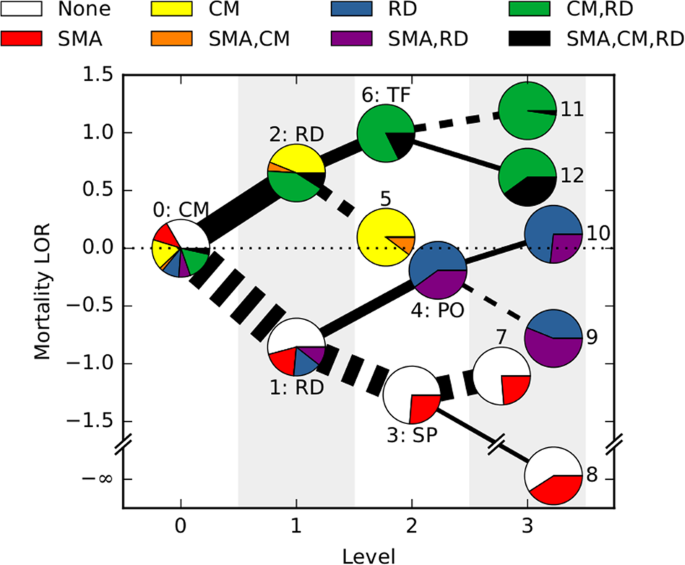
We first asked which features were most strongly predictive of patient outcome. We used a statistic called mutual information to provide a “decision tree” for individual patient risks, rather than a simple yes/no output. For example, if a patient does not have the cerebral form of malaria, the next most important question is whether they experience respiratory distress. If so, the next important question is whether their posture is abnormal. Following these branches of the decision tree, the overall risk of mortality can be determined for an individual, specific patient.
We next asked how malaria progresses in individual patients. We used an algorithm called HyperTraPS (hypercubic transition path sampling) to produce the first “roadmap” of malaria progression, providing the probability with which each symptom is acquired at each possible stage of disease. HyperTraPS can learn these dynamic disease pathways using just a collection of single-time “snapshots” of patient symptoms. We compared these detailed pathway predictions with a new survey of malarial experts and found a good agreement with their responses. We then identified those features that best distinguished pathways that lead to patient survival from those that lead to patient death. We used these features to produce a new and detailed classifier of mortality risk for individual patients. This successfully assigned independent patients to low and high risk groups, notably with a low false negative rate (few high-risk patients were classified as low-risk).
We hope that this joint analysis, using a decision-tree approach and a risk classifier based on disease progression, is valuable in refining the current broad-strokes picture of severe malaria risk. We also believe that this mathematical framework to harness large, but incomplete and mixed, datasets will find use in the study of other progressive diseases.
Follow the Topic
-
npj Digital Medicine

An online open-access journal dedicated to publishing research in all aspects of digital medicine, including the clinical application and implementation of digital and mobile technologies, virtual healthcare, and novel applications of artificial intelligence and informatics.
Related Collections
With Collections, you can get published faster and increase your visibility.
Digital Health Equity and Access
Publishing Model: Open Access
Deadline: Mar 03, 2026
Evaluating the Real-World Clinical Performance of AI
Publishing Model: Open Access
Deadline: Jun 03, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in