A hidden hitchhiker brought to light - on how we isolated a novel virophage
Published in Microbiology

Back in 2018 the Beja lab was divided into two main research teams, one working on microbial rhodopsins, while a second focused on Auxiliary Metabolic Genes on marine phages. I decided to bridge between those groups and study a viral rhodopsin that is probably being used as an auxiliary metabolic gene by a giant virus infecting the abundant marine algae Phaeocystis globosa. A few months later I successfully set up this virus-host system in the lab, and after two years of work I had to accept that the viral rhodopsin was nowhere to be seen. Although it was being expressed, we could not detect the protein by any means (and we tried many: proteomics, targeted proteomics, antibodies, activity assays…). We were frustrated and discouraged, and started to question the very basic foundation of the project: Do we have the right virus? We sent our lysate for sequencing. The giant virus was exactly the one we expected it to be, but we found a surprising hitchhiker in our sample.

A second small viral genome was assembled from our lysate, and we got ecstatic! A hidden player! What a treasure! A quick literature search brought us back to the ground, as we realized that it was very similar to the Phaeocystis globosa virus virophage genome discovered in 2013 by our colleagues in the Brussaard lab, for whom no viral particles were ever detected. Nonetheless, we were curious about this little virus who was a silent passenger in the lysate for many years. We were also lucky to be a little ignorant. Had we known any better, we would have not embarked on the quest to isolate it, for all similar viruses discovered to date, the Polinton-like virus group, were reported solely as genomic fragments.
Some PCRs, SYBR-staining and TEM imaging later we were ecstatic again, as our small virus was indeed a separate entity, a dsDNA genome packed in its own small capsid. This virus is also proven to be a virophage, a virus that parasitizes on virocells- eukaryotic cells actively infected by viruses, rather than directly infecting a eukaryotic host. To this date virophages were only classified within the Ladaviridae family (like Sputnik or Mavirus), however our virus is not related to any of them. It took us a while to realize the importance of this finding: “Virophage” is a lifestyle, a reproduction strategy, independently developed by different viral families. We named our virus “Gezel”. Gezel is a Dutch word assigned to a pupil who was trained in some profession and to become an expert he joined his master, and typically lived in the same household. In Hebrew “gezel” means “theft”, a suitable name for this virus, which uses its viral host resources to reproduce within the eukaryotic cell.
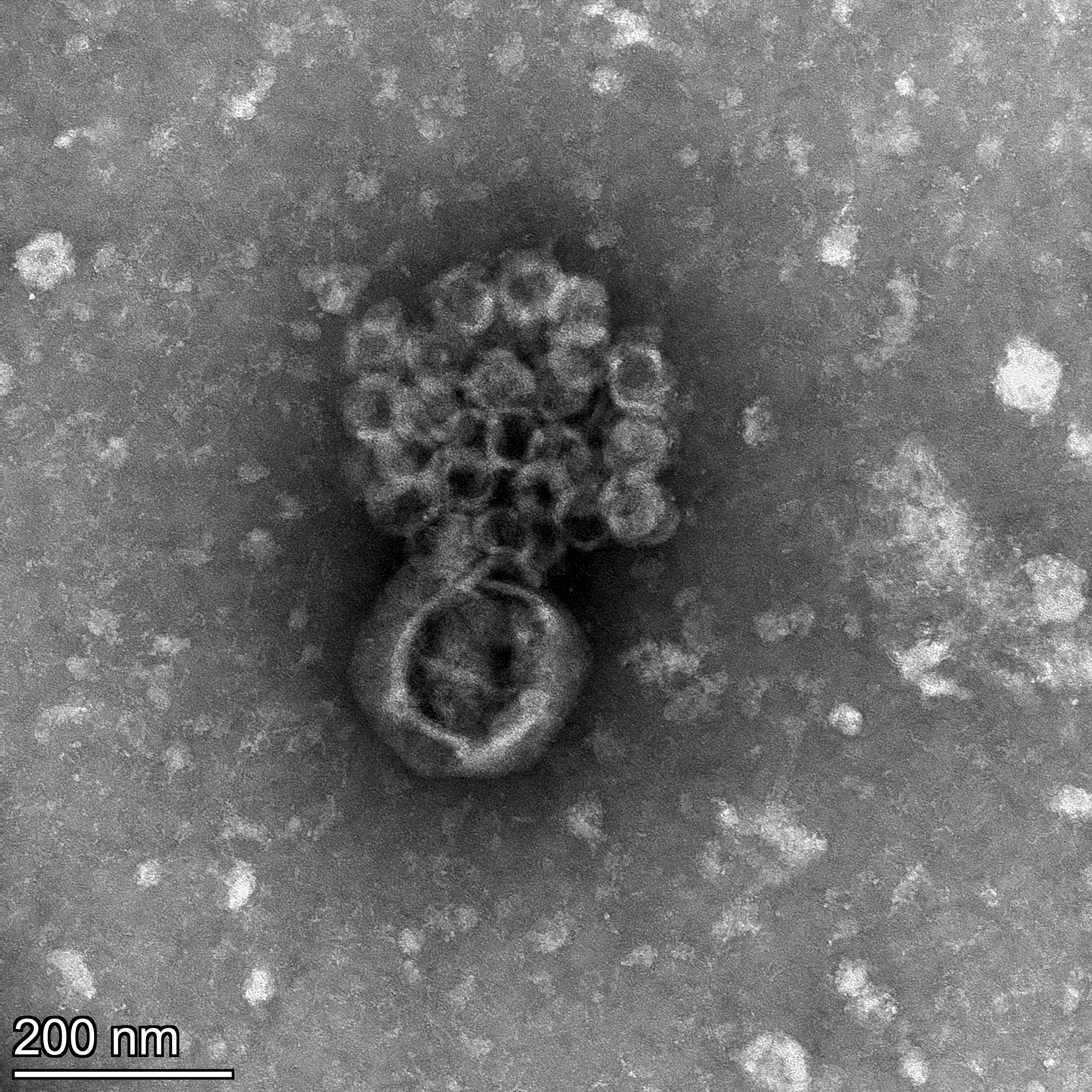
A quick search in the algal host genome showed various viral-like elements similar to Gezel,
meaning that this virus is capable of integrating into the cellular host’s genome. This double life-style suggests Polinton-like viruses are in the gray area between transposable genetic elements and viruses. It was very exciting to unveil the layers of parasitism in this system: the algae is being parasitized by both viruses, by direct infection and integration to its genome. Gezel prays on the giant virus, impairing its reproduction. Surprisingly, Gezel itself is a victim of parasitism. An intronless site-specific homing endonuclease located itself between its minor and major capsid proteins. This selfish genetic element might hitchhike on Gezel, but it could also serve as a defense mechanism against other virophages, although we did not see it expressed in our experiments, contrary to all other Gezel ORFs.

Finally, we propose a framework to classify these viruses within the virosphere, based on phylogeny and gene content. To date the classification of these elusive viruses was based on the distinction between Polintons (first classified as large transposons, currently considered integrated viruses), Polinton-like viruses (integrated or free viral-like genomes) and virophages (classified in the Lavidaviridae family of viruses). Gezel blurs these distinctions for it is a free-living virus, can integrate into the algal genome and leads a virophage lifestyle. Therefore, we pose that a revision of these taxonomic designations is imperative. Gezel-like viruses share a set of genes and cluster in a single well supported clade, thus we propose they should be classified together in a new viral genus Gezelvirus, with the new species Gezelvirus phaeocystis to accommodate this first isolate.
Overall this project felt like a rollercoaster. I failed on my initial purpose of merging both research topics in the lab, and worse, I ended up creating a third research direction. However, it was an exciting project to work on, shedding new light on the origins of viruses and parasitism.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in