A Secret Weapon in Plant Infection: How Oomycetes Use Carbohydrate Oxidation to Evade Plant Immunity
Published in Sustainability, Cell & Molecular Biology, and Plant Science
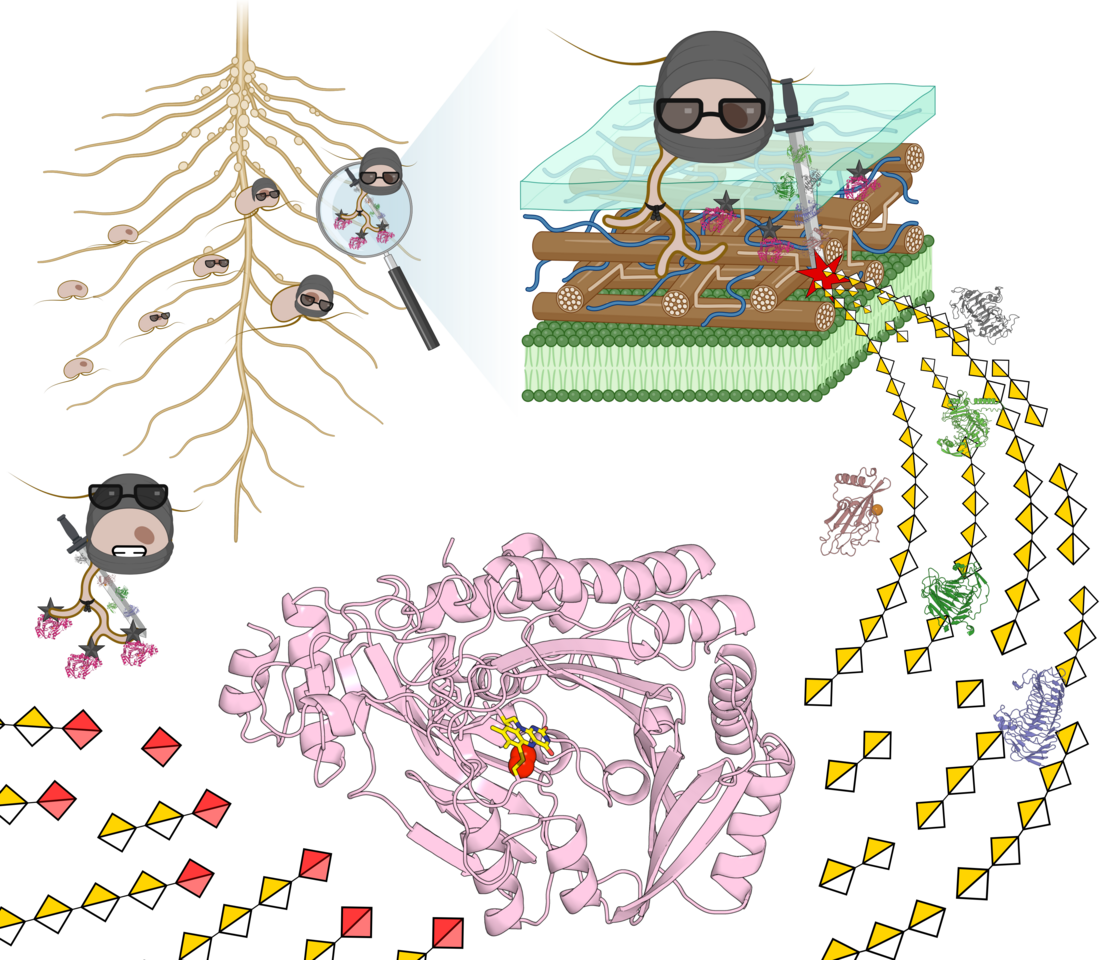
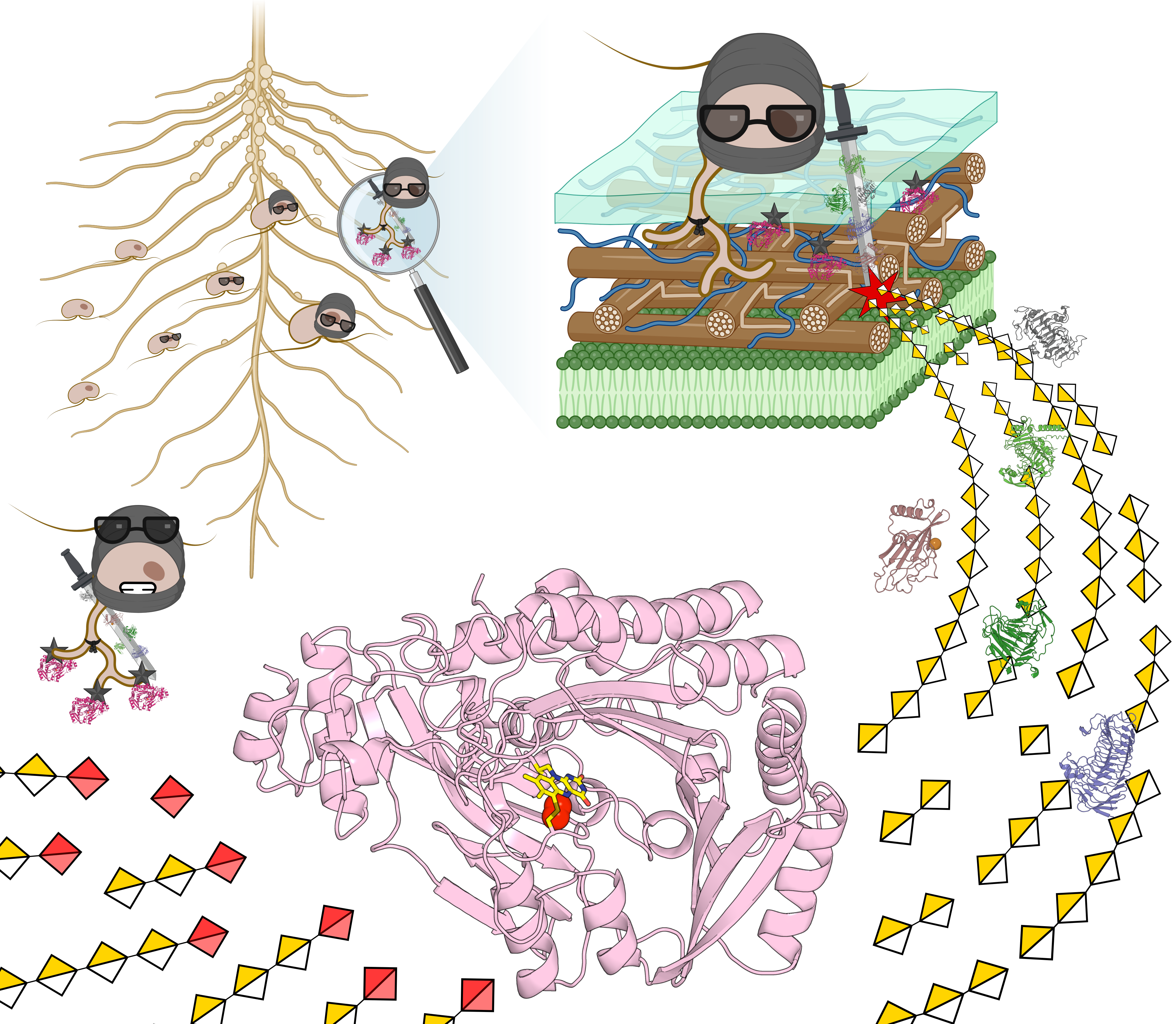
What would you think if someone told you that ninjas and water moulds have something in common? It might sound far-fetched, but it’s the kind of idea that kept running through my mind while we were piecing together our work during my PhD project.
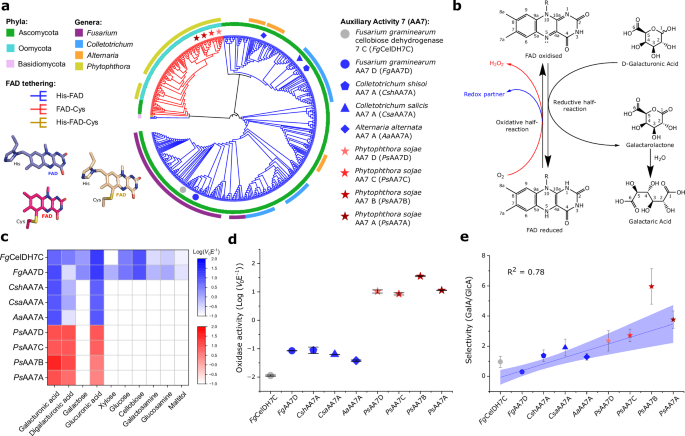
The journey to reach such an idea started back in 2021 and builds on foundational work by my PhD supervisor Prof. Maher Abou Hachem (DTU, Denmark). His collaboration with Senior Scientist Jean-Guy Berrin (INRAE, France) led to the discovery of a peculiar FAD-dependent oligosaccharide-oxidising enzyme from the cereal pathogen Fusarium graminearum, classified in the CAZy database under the Auxiliary Activity family 7 (AA7)1. Its unique structure and activity profile holds significant biotechnological potential for lignocellulosic waste bioconversion, sparking renewed interest in this family of enzymes. Yet one broader question puzzled us—beyond their industrial relevance, why do AA7 enzymes appear to be so widely expressed across fungi, while they occur conspicuously in infamous plant pathogenic water moulds?
Therefore, at the start of my PhD, we began exploring the AA7 clade that harboured our previously reported enzyme from F. graminearum1. Interestingly, this entire clade was glowing with intriguing features. Most notably, alongside numerous fungal plant-pathogen enzymes, we identified an atypical branch of Oomycota sequences enriched in 18 Phytophthora species. Why was this exciting? Oomycota are a distinct phylum of water moulds including some of the most devastating plant pathogens—particularly species from the Phytophthora genus2. These pathogens are infamous for causing the Great Irish Famine2 and for continuing to inflict major crop losses worldwide3. This conspicuous conservation of Phytophthora AA7 sequences within the same clade as notorious fungal plant pathogens was a magnet for our curiosity, hinting at an important shared biological role.
Therefore, alongside four fungal representatives, we selected four homologues from Phytophthora sojae for further biochemical characterisation. Why P. sojae? Because, beyond being a devastating soybean pathogen responsible for an estimated 2 billion $ in annual losses worldwide, it is widely used as a model species in Phytophthora biology4. With the collaboration of NMR expert Prof. Sebastian Meier (DTU, Denmark), and support from our collaborators at INRAE (France), we achieved an in-depth characterisation of these new AA7 enzymes. In parallel, through the collaboration with Prof. Jens Preben Morth (DTU, Denmark) and the work of the former postdoc in our lab Dr. Sanchari Banerjee (DTU, Denmark), we solved the crystal structures of two AA7 enzymes, including one from P. sojae. This cross-disciplinary work reinforced the uniqueness of this AA7 clade, revealing for the first time a 6-S-cysteinyl-FAD architecture. Initially we were happy to have discovered the first secreted microbial FAD-dependent enzymes capable of oxidising galacturonic acid. But the real thrill came about when we demonstrated that the Phytophthora enzymes were uniquely able to oxidise oligogalacturonides (OGs), derived from pectin—the main constituent of the middle lamella that joins plant cells together. We showed that these enzymes are active on the OGs consisting of 16 units of galacturonic acid. Some readers might say, 'Big deal!' However, our excitement was spurred by the known potent elicitor activity of OGs of this size. For example, OGs released by pectin degradation during pathogen invasion are detected by plant receptors as damage associated molecular patterns (DAMPs), which alert the plant to the ongoing pathogen attack, triggering defensive immune responses5. This is a crucial defence mechanism, given that plants lack adaptive immunity and instead rely on a ‘sentinel’ system to detect and respond to danger signals. Through discussions with Dr. Jean-Guy Berrin, we started a collaboration with the Phytophthora expert Prof. Suomeng Dong (Nanjing Agricultural University, China). With his team, we mapped the expression of pectin-active enzymes (pectinozome) across different infection stages. Strikingly, distinct genes encoding our newly discovered OG oxidases were highly co-expressed with counterparts conferring the degradation of the pectin homogalacturonan during early infection, hinting at an important role in the early infection stages. The team of Prof. Dong performed key experiments using their state-of-the-art CRISPR-based technology to knock out (KO) single OG oxidase genes. The phenotype of the KO strains was evaluated in a soybean infection model, demonstrating a significant decrease in pathogen burden. These findings, combined with the early-stage expression during infection, led us to hypothesise that the OG oxidases impair the recognition of OGs by infected plants. By oxidising OGs, P. sojae disrupts the plant’s early warning system, effectively ‘blinding’ it by disabling OG-mediated immune responses and thereby facilitating a more efficient infection. Doesn’t this sound like a stealth manoeuvre straight out of a ninja movie?
Beyond deepening our understanding of Phytophthora infection strategies, this highly cross-disciplinary work provides the first evidence of an in vivo role for AA7 enzymes. Ultimately, this discovery not only advances our understanding of plant-pathogen virulence mechanisms but also opens our eyes to a novel virulence strategy. Further work holds promise to unlock innovative approaches for crop protection.
References.
- Haddad Momeni, M. et al. Discovery of fungal oligosaccharide-oxidising flavo-enzymes with previously unknown substrates, redox-activity profiles and interplay with LPMOs. Nat Commun 12, 2132 (2021).
- Brasier, C., Scanu, B., Cooke, D. & Jung, T. Phytophthora: an ancient, historic, biologically and structurally cohesive and evolutionarily successful generic concept in need of preservation. IMA Fungus 13, 12 (2022).
- Fones, H. N. et al. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat Food 1, 332–342 (2020).
- Tyler, B. M. Phytophthora sojae : root rot pathogen of soybean and model oomycete. Molecular Plant Pathology 8, 1–8 (2007).
- Pontiggia, D., Benedetti, M., Costantini, S., De Lorenzo, G. & Cervone, F. Dampening the DAMPs: How Plants Maintain the Homeostasis of Cell Wall Molecular Patterns and Avoid Hyper-Immunity. Front. Plant Sci. 11, 613259 (2020).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
What are SDG Topics?
An introduction to Sustainable Development Goals (SDGs) Topics and their role in highlighting sustainable development research.
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in