Adapting to resistance
Published in Ecology & Evolution
Bacteria can become resistant to antibiotics in a whole bunch of ways. One really fast and effective way is gaining an extra bit of DNA, called a plasmid, from other bacteria. The plasmid provides bacteria with the genes needed to become resistant to specific antibiotics. Resistance can spread between bacteria (even of vastly unrelated species) through the exchange of plasmids, but is the gaining of a resistance plasmid all the cell has to do to deal with antibiotics, or is something more complicated happening?
I set out to see what would happen to the bacteria (E. coli in this case) and the plasmid if they were exposed to antibiotics for a long time, enough so that the bacteria couldn’t grow without the plasmid. The plasmid I was interested in provided resistance to two different commonly used antibiotics: ampicillin and tetracycline. I knew that both antibiotics and both resistances work in very different ways. But, would exposure to these two antibiotics cause different evolutionary paths to be followed by the bacteria?
To test this I used a technique called experimental evolution: bacterial populations containing the resistance plasmid were grown in different antibiotic environments and allowed to evolve for 500 generations. To test what affect the antibiotics were having we had a range of treatments including no antibiotic, mono- and combination-therapies of tetracycline and ampicillin. This was only the start of the experiment, now I had to figure out what, if anything was had happening to my bacteria because of the antibiotics.
I stumbled upon my first discovery by chance. The first thing I wanted to do was isolate any changes that had happened in the bacteria’s DNA from the plasmid DNA. This required removing the plasmid, which had shared the company of the bacteria for all that time. One step of this required me to remove some of the bacteria by adding a third antibiotic, also commonly used, called chloramphenicol. Looking at the negative controls – experiments expected to produce nothing as a check on the method – I noticed that the bacteria that had evolved in the presence of tetracycline (but not ampicillin) were growing on the chloramphenicol plates despite never being exposed to this antibiotic before. At first this set off alarm bells, growth on the negative controls is not good! But after a bit of digging I realised that the bacteria grown in the presence of tetracycline had developed mutations on their chromosome that allowed them to resist ampicillin, tetracycline and chloramphenicol to a low level without the help of the resistance plasmid.
Next, I wanted to see if the resistances on the plasmid had changed at all, so I put the evolved plasmid that had been exposed to a load of antibiotic for a long time, into my ancestral bacteria, which had not. To make sure I got the plasmid into the ancestral bacteria I gave them some more tetracycline (enough that the bacteria couldn’t grow without the plasmid). I noticed that the bacteria that had evolved in the presence of tetracycline really didn’t look very healthy despite having a plasmid that I thought should protect them. After further experiments, I realised that mutations had arisen on the plasmid in the genes that provide resistance making it less resistant but having a more comfortable relationship with the bacteria.
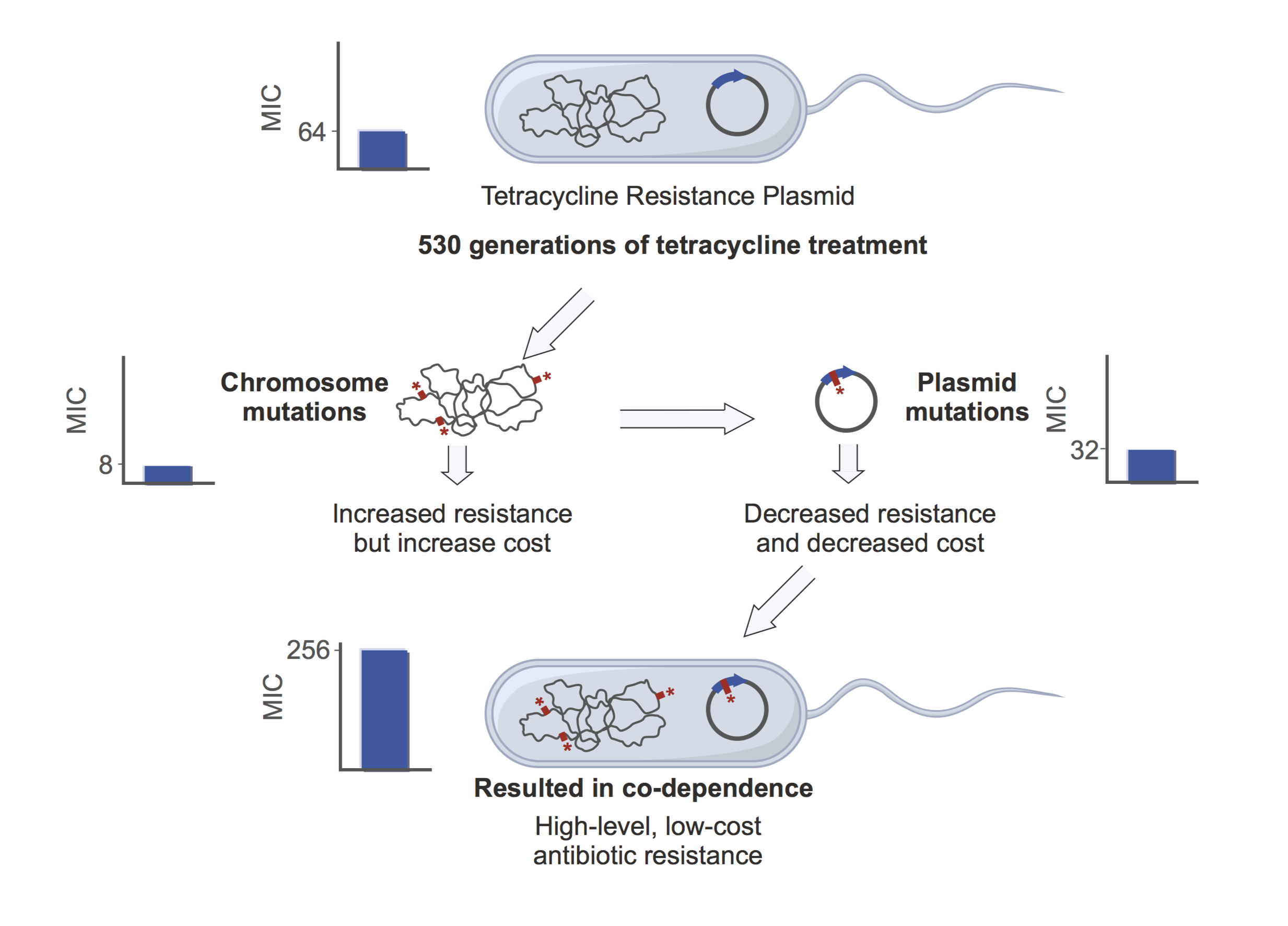
I figured out that something really quite counterintuitive was happening. During the 80 days the bacteria were exposed to tetracycline, first they gained additional resistance mutations on the chromosome, but this meant that the plasmid was now somewhat redundant. The level of resistance provided by the plasmid could be tuned down, through specific mutations, which also made it less costly. This produced a plasmid and host that were now dependent upon each other when exposed to this antibiotic. A positive side affect (at least for us) of this coadaptation between the plasmid and host is that the plasmid is actually less effective in other bacterial hosts, and is now less likely to spread to other bacteria in the future. On the down side the bacteria itself has become more resistant to this and other antibiotics.
Gaining resistance plasmids is just the start of the bacteria’s journey to become resistant; the marriage between plasmid and bacteria is a complex one, involving both compromise and changes in behaviour. It is a relationship we need to unpick further in order to best preserve the antibiotics we have for use in both critical and routine medical procedures.
Our paper in Nature Ecology and Evolution can be found here: http://go.nature.com/2tFSxxU





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
1.hello, that other mechanisms influence the resistance and that they form are translated into the bacterial genome. Thank you