After the Paper | Utilizing chemical mutagenesis to study the impact of mutations on gene expression and alternative splicing
Published in Ecology & Evolution

In October 2021, I reached a significant milestone of my graduate school journey by publishing my first first-author paper titled: The genome-wide rate and spectrum of EMS-induced heritable mutations in the microcrustacean Daphnia: on the prospect of forward genetics in Heredity. The primary objective of this project was to design a forward genetic screening approach in the freshwater microcrustacean Daphnia, aimed at facilitating the study of gene function. Forward genetic screening approaches have been developed and successfully implemented in gene function discovery studies for a variety of model organisms including Caenorhabditis elegans (Brenner 1974), Arabidopsis thaliana (McCallum et al. 2000) and Drosophila melanogaster (Lewis and Bacher 1968). However, such an approach was still lacking for Daphnia, despite its emergence as a promising model organism in the field of Evolutionary Biology.
As a first step towards developing our forward genetic screening approach, we devised a mutagenesis protocol using the widely employed chemical mutagen ethyl methanesulfonate (EMS) to generate our Daphnia mutants (Figure 1A). Sexually mature Daphnia females were subjected to varying concentrations of EMS (e.g., 10mM, 25mM, 50mM, and 100mM) for 4 hours, with the 10mM and 25mM concentrations showing the highest survival rates. Next, we employed whole-genome sequencing to examine and characterize the mutation rate and spectrum in mutants exposed to these two EMS concentrations.
Our findings demonstrated that both EMS treatments successfully induced mutations in the Daphnia genome. The 10mM treated mutants exhibited an average base substitution rate of 1.17 × 10−6 per site per generation, while the 25mM treated mutants had an average rate of 1.75 × 10−6 per site per generation (Figure 1B). These rates were significantly higher than the spontaneous mutation rate of 2.30 × 10−9 and 7.17 × 10−9 per site per generation, respectively (Flynn et al. 2017, Keith et al. 2016). The mutation spectrum for both EMS treatment conditions were dominated by GC to AT transitions, averaging 87% and 86% for the 10mM and 25mM lines respectively (Figure 1C). This represents a significant rise compared to the approximately 66% GC to AT ratio documented in Daphnia spontaneous mutation accumulation lines (Keith et al. 2016). Furthermore, we showed that EMS mutagenized females (F0) produce unique mutant offspring (F1) for at least 3 generations, indicating that each oocyte was individually impacted and harbored unique mutations. Additionally, the mutant offspring showed a mutation rate and spectrum similar to that of the F0 individuals (Figure 1 B, C).
In light of our results, we conducted a power analysis to provide guidance for genetic screening efforts. We estimated that for all genes in the Daphnia genome to be mutated at least once with 95% probability, 750 F1s need to be screened. For mutations to be obtained in the homozygous state with 70-80% probability, we recommend that 4-5 F2s be collected from each F1 mutant.
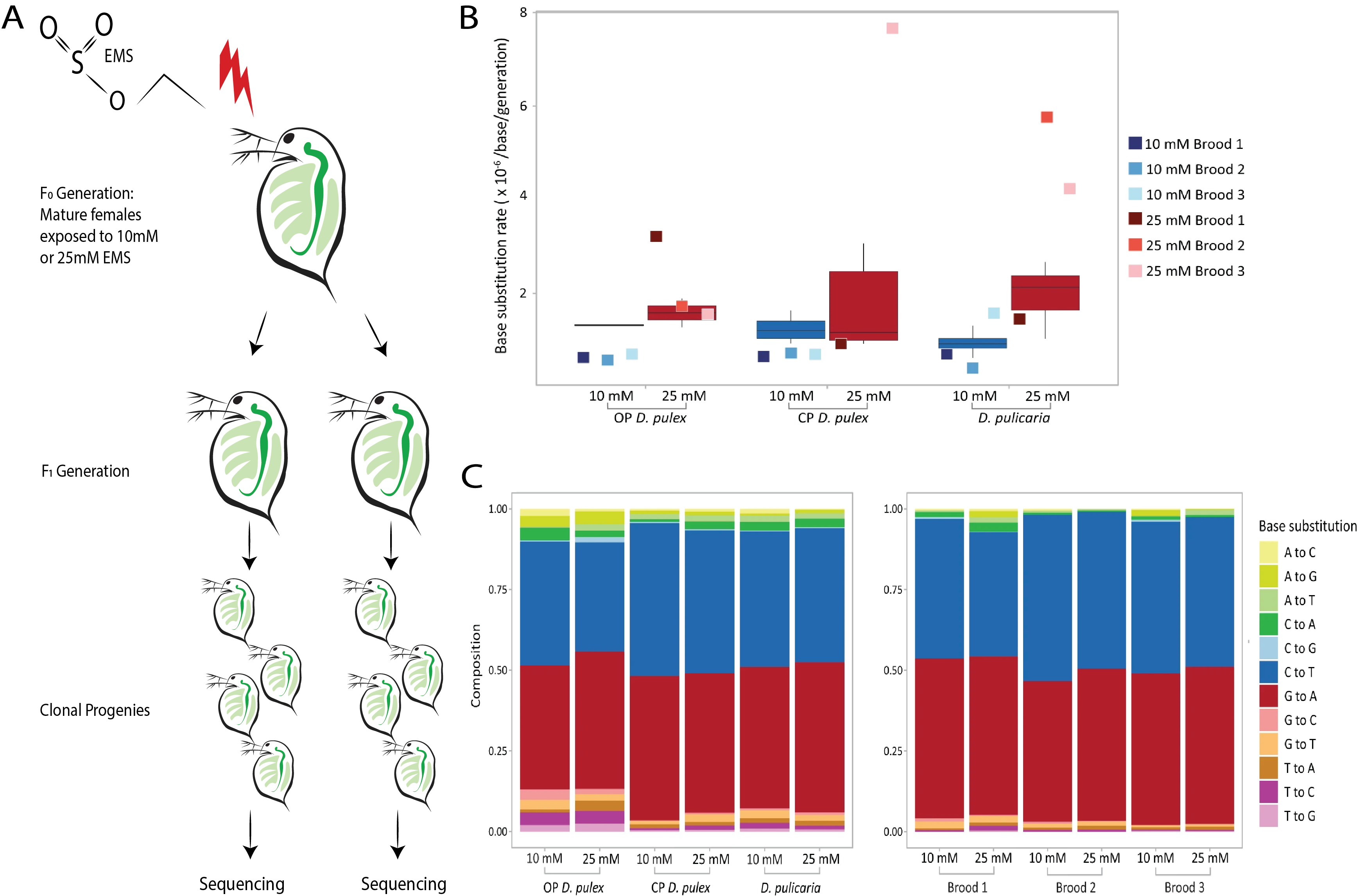
Figure 1. (A) Mutagenesis protocol using ethyl methanesulfonate. (B) Base substitution rate per generation across different species exposed to 10mM and 25mM EMS concentrations. (C) Proportion of base substitution across species and broods.
To build upon this work we recently published a follow up study titled: The effects of mutations on gene expression and alternative splicing in Proceedings of the Royal Society B, where we estimated the impact of EMS-induced mutations on whole-genome gene expression and alternative splicing patterns in Daphnia mutants. Furthermore, we determined the contribution of cis- and trans-mutations to differential gene expression and alternative splicing. Our results showed that the variance in gene expression and alternative splicing between wild-type and EMS mutant lines are mainly due to the pleiotropic effects of trans-mutations. Cis-mutations impacted a relatively smaller number of genes and gene expression was not always affected. Lastly, we showed that differentially expressed genes were significantly associated with exonic mutations, highlighting the importance of exonic mutations as a driver of altered gene expression.
References
1. Brenner S (1974) The genetics of Caenorhabditis Elegans. Genetics 77:71–94
2. Flynn JM, Chain FJJ, Schoen DJ, Cristescu ME (2017) Spontaneous Mutation Accumulation in Daphnia pulex in Selection-Free vs. Competitive Environments. Molecular Biology and Evolution, 34(1), 160–173.
3. Keith N, Tucker AE, Jackson CE, Sung W, Lucas Lledó JI, Schrider DR, Schaack S et al. (2016) Generation of loss- or gain of function mutants and weak nonlethal alleles. Genome Research, 26(1), 60–69.
4. Lewis EB, Bacher F (1968) Method of feeding ethylmethane sulfonate (EMS) to Drosophila males. Drosophila Information Service 43:193
5. McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeted screening for induced mutations. Nat Biotechnol 18:455–457


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in