Anti-cancer therapy is associated with long term epigenomic changes in childhood cancer survivors
Published in Cancer
Main findings
In our study, we found a novel set of epigenetic changes that could be detected immediately following anti-cancer therapy in childhood cancer patients and were also detected many years after therapy in adult survivors of childhood cancer, including once loci significantly associated with premature death. These could function as biomarkers for predicting late health effects in childhood cancer survivors and also for assessing the likely impact of novel therapies and altered treatment protocols on long-term outcomes.
What is the relevant background?
Cancer treatments have drastically improved survival rates of childhood cancers, but they also substantially increase the risk of long-term chronic health conditions among survivors (1,2). Common morbidities include cardiovascular diseases, diabetes mellitus, obesity, subsequent cancers, and premature ageing.
Whilst the risk of morbidities and early mortality in cancer survivors is well-established, there is limited understanding of the underlying mechanisms. Epigenetic mechanisms are plausible candidates mediating these effects, as the changes are modifiable in response to internal and environmental factors and epigenetic changes in early life have been linked to increased susceptibility to disease in adulthood, (3). The best-characterized epigenetic modification is DNA methylation, which involves the addition of methyl groups on cytosine bases next to guanine (CpG), and is a key regulator of gene expression. Altered DNA methylation patterns and age associated changes in DNA methylation have been associated with the development of multiple chronic diseases and reduced life expectancy (4).
What did we investigate?
We investigated the hypothesis that anti-cancer therapies induce long-term DNA methylation changes which ultimately contribute to adverse late health effects. To do this, we conducted epigenome-wide association studies (EWAS) to identify the regions of DNA methylation that were altered following anti-cancer treatment in childhood cancer patients. To see if these changes were evident in long-term survivors, we investigated these regions in a different group of adult survivors of childhood cancer many years post-treatment. We also looked at markers of epigenetic ageing using Horvath’s DNA methylation clock in childhood and adult cancer survivors (5). Finally, we investigated if the differentially methylated regions were associated with later-life adverse health outcomes.
What did we find in the childhood cancer patient samples?
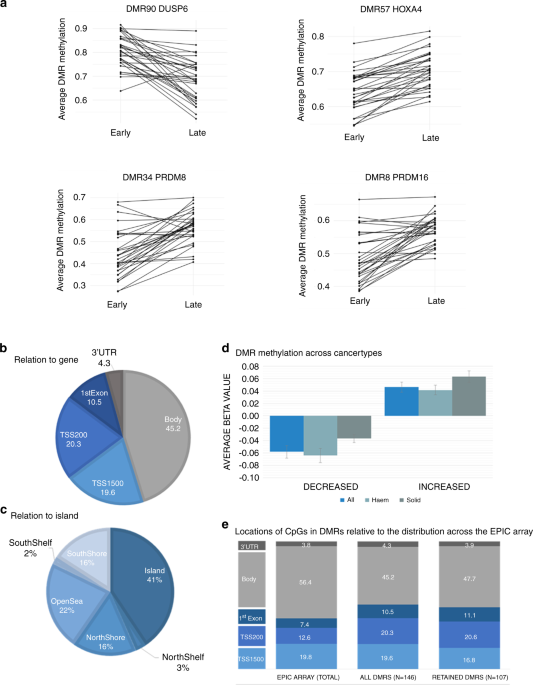
We used matched samples (before and after treatment) from childhood cancer patients mainly undergoing treatment for haematological cancers. There were 146 differentially methylated regions that were consistently altered post-treatment. Most of these regions had increased methylation and were found in or close to promoter regions, which could influence gene expression. There was no evidence of accelerated ageing at the end of treatment.
What did we find in the adult survivors?
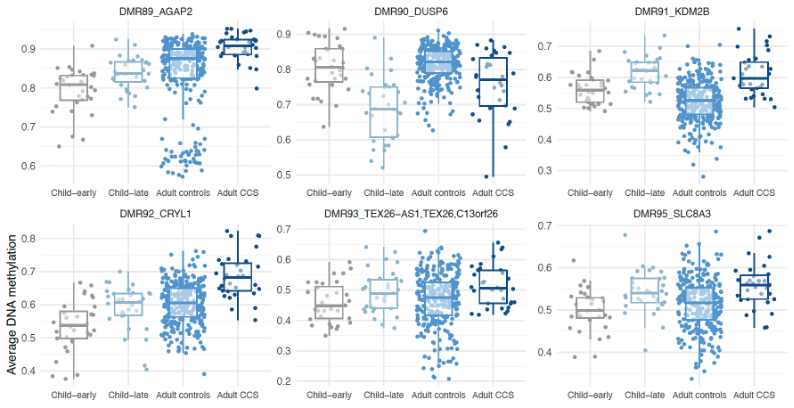
To investigate whether these DNA methylation changes were transient or maintained into adulthood, we examined DNA methylation patterns in survivors of childhood/young adult cancer compared to an age and sex-matched control population. We collaborated with researchers with data from the European Prospective Investigation into Cancer and Nutrition (EPIC) study; a large and extensively phenotyped, longitudinal cohort. Individuals with previous diagnoses of cancer (before age 25) who were cancer-free for at least 10 years at baseline were included. 73% of the regions (107 of the 146) identified in the childhood patients were also differentially methylated in the adult cancer survivors. This suggests that DNA methylation changes which occur following treatment for cancer in childhood may be stable and still present many years after treatment.
The consistency of the changes was surprising considering there were different types of cancers and likely many different types of treatment protocols
In contrast to the childhood samples, the adult survivor population showed clear evidence of accelerated epigenetic ageing (average of 5.5 years older years than the control cohort). Adult cancer survivors had higher rates of early mortality and second cancers as we might expect, but those who had premature mortality had slightly higher levels of DNA methylation overall, and significantly lower levels of methylation in the DUSP6 gene.
Why these findings are important
We identified a common set of methylation changes that were present across different childhood cancers following exposure to anti-cancer therapy, which could be useful as biomarkers in future studies of childhood cancer survivors. Molecular markers for predicting late effects are of significant clinical utility, both in risk prediction to enable targeted follow-up care for those at greatest risk of significant late effects, and for monitoring the likely long-term health impacts of novel therapies or altered treatment protocols. Mechanistic studies may reveal more insight as to whether these sites could also be therapeutic targets. As childhood cancer is a rare disease, additional large studies are required to further validate and replicate these findings.
- Bhatia S., et al. Collaborative Research in Childhood Cancer Survivorship: The Current Landscape. J Clin Oncol 2015, 33, 3055-3064.
- Reulen R.C., et al. Long-term risks of subsequent primary neoplasms among survivors of childhood cancer. JAMA 2011, 305, 2311-2319.
- Relton C.L. & Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. Plos Med 2010. 7(10):e1000356.
- Fraga M.F. & Esteller, M. Epigenetics and aging: the targets and the marks. Trends in Genetics 2007, 23, 413-418.
- Horvath S. & Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018, 19, 371–384.
Follow the Topic
-
British Journal of Cancer

This journal is devoted to publishing cutting edge discovery, translational and clinical cancer research across the broad spectrum of oncology.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in