Artificial Intelligence in Multiple Myeloma: towards personalized risk stratification and treatment selection
Published in Cancer

Multiple Myeloma (MM) is a frequent and mostly incurable disease of the bone marrow which has been the focus of multiple new drugs in the last years. Despite numerous advances in our comprehension about the biology of MM, risk stratification and upfront treatment selection are still mostly based on clinical and cytogenetics data, with suboptimal results. Our limited comprehension about the mechanisms of drug response poses a pitfall for novel drug development, and probably underlies some of the recent clinical trial failures in the field.
Machine learning is being actively applied to select optimal investments in the fintech industry, and to guide client-oriented marketing in the retail sector. We reasoned that such excellent experiences in so diverse fields proved that predicting key outcomes in complex systems is within our reach. Indeed, growing evidence indicates that the application of artificial intelligence in cancer dramatically improves disease prognostication. After all, cancer is nothing but a complex multidimensional system, isn’t it?.
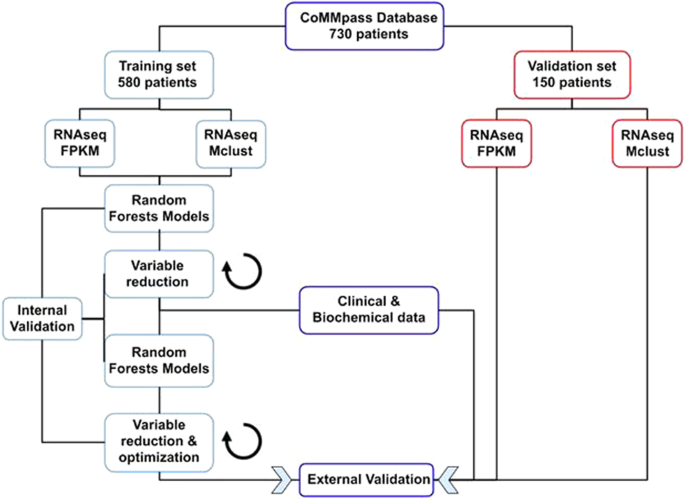
Therefore, we believed that such an approach could be used to close the gap between our comprehension of molecular biology and clinical decision making in MM. With this in mind, we struggled to develop a model of survival based on clinical, biochemical and molecular data that could be used to individualize risk stratification and guide upfront treatment selection. Using data produced by the CoMMpass consortium, we developed a 50-variable random forests model named Iacobus-50 (IAC-50) that predicted overall survival with high concordance between both training and validation sets. This model included the following covariates: patient age, ISS stage, serum B2-microglobulin, first line treatment and the expression of 46 genes. Survival predictions for each patient considering the first line of treatment evidenced that those individuals treated with the best predicted drug combination were significantly less likely to die than patients treated with other schemes. This was particularly important among patients treated with a triplet combination including bortezomib, an immunomodulatory drug (ImiD) and dexamethasone. Finally, the model showed a trend to retain its predictive value in patients with high-risk cytogenetics.
The novelty of IAC-50 is that it follows a top-down approach where patient outcomes are at the core of the model, instead of relying on imperfect patient subgrouping. By bringing together artificial intelligence and genomic medicine, we believe that our results are the tipping point to start the à la carte treatment revolution in the field of MM.
Follow the Topic
-
Leukemia

This journal publishes high quality, peer reviewed research that covers all aspects of the research and treatment of leukemia and allied diseases. Topics of interest include oncogenes, growth factors, stem cells, leukemia genomics, cell cycle, signal transduction and molecular targets for therapy.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in