Automated removal of non-homologous sequence stretches in phylogenomic datasets
Published in Ecology & Evolution
More often than not, phylogenomic datasets contain errors, different kinds of errors. These can be for example the mixture of paralogous gene copies, undetected contamination or cases of lateral gene transfers. Because these errors confound the genuine phylogenetic signal and represent clear violation to the principles of phylogenetics, phylogenomicists track them down like pest and most are cleaned away (although not always).
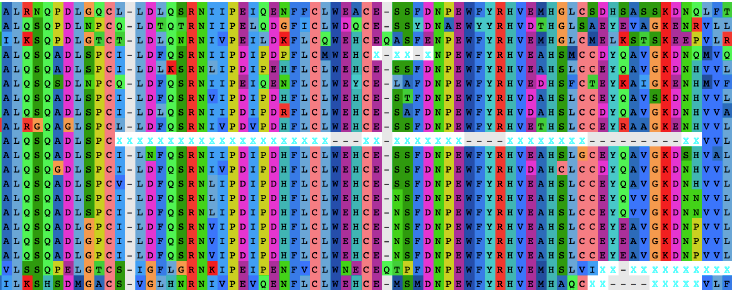
Another kind of errors, often harder to detect and less talked about, remain a rampant issue in phylogenomics: the non-homology of character stretches in individual sequences. Anyone who has assembled or inspected a phylogenomic dataset will recall seeing amino acid stretches that look out of place, like if they don’t belong to the sequence or were misaligned. This becomes obvious after direct visualization of a few gene alignments. A typical case scenario for such fragments might be produced by sequencing errors that insert (or delete) a nucleotide, leading to the disruption of the reading frame in protein-coding genes when these are translated into amino acid sequences. With poor sequence quality, it is not uncommon that the reading frame is re-established by a second deletion (or insertion) downstream, producing stretches of non-homologous amino acids within an otherwise legitimate sequence. Another common case would be the use of poorly annotated genes extracted from a genome, resulting for example from poor prediction of intron-exon boundaries. Upon multiple sequence alignment, this non-homologous region might be forced into the alignment producing wrong homology inferences. As an example, look at the top panel of figure below, which contains sequences from the beginning of the Hedgehog-interacted protein (HHIP) obtained from full vertebrate genomes at OrthoDB (https://www.orthodb.org/?level=&species=&query=EOG090702V5).

So the issue here is how does one detect these regions? As it turns out, this is not an easy problem and there was no easy fix (beyond manual curation, which is impractical for more than a few genes and is of course highly subjective). Also, the easier alternative of simply ignoring these types of errors is not justified, because experience shows that they are a widespread occurrence in phylogenomic datasets and thus likely to confound the phylogenetic signal, especially when this signal is weak to start with. Note that the task at hand here is conceptually different than removing ambiguously aligned sites from an alignment (for which several tools exist, e.g. trimal, gblocks, bmge); here, we deal with non-homologous residues (amino acids) in individual sequences, which are not necessarily associated with regions of poor alignment but should be excluded anyway.
This issue was brought up around a cup of tea on a wintry Sunday afternoon, during an impromptu discussion between the authors of what will become a new tool to handle non-homologous characters in protein sequences. In some ways, this was a lucky circumstance with a happy ending, or what can happen when a statistician and an empirical phylogenomicist have tea together… The new tool in question was called PREQUAL, to reflect on what it does: it cleans sequences, it can and should be implemented early in phylogenomic pipelines, and improves the quality of datasets. We think it fills an important gap in phylogenomics, by automatically handling stretches of non-homologous characters in sets of (unaligned) sequences. These regions share no statistical support for homology with the residues in any other sequence in the set. Under the hood, PREQUAL uses a full probabilistic approach (pair hidden Markov models): it calculates posterior probabilities to evaluate the evidence for homology of each character in each sequence; residues with insufficient evidence of homology are masked or removed.
PREQUAL is very user-friendly, with easy installation and usage that requires minimal user intervention. The default values should work well for most phylogenomic datasets, but it is possible to fine-tune the parameters to fit the needs of every user. PREQUAL is designed for amino acid sequences, although it can also handle protein-coding DNA sequences. PREQUAL is fast, taking only 1 minute to process a set of 90 sequences 700 amino acid long. Therefore, pre-alignment masking does not impose an additional computational burden and can be easily incorporated into existing bioinformatic pipelines.
But how do we know that PREQUAL works? That is, does it remove the incriminated amino acids and only those ones? To answer this question, we benchmarked PREQUAL using simulated error-free gene sequences, which we then corrupted by including frameshifts and annotation errors at known (random) positions. From these simulation experiments, we see that across all tested conditions PREQUAL showed a high accuracy (proportion of correctly identified true and erroneous residues). But of course the next question was: are these simulations representative of what real data look like? This isn’t trivial! We tested PREQUAL on several published phylogenomic datasets and carefully analysed results, which suggested that PREQUAL does a pretty good job at classifying residues as correct or erroneous. Based on our experience with empirical datasets, PREQUAL does a similar job from that of an expert eye, if we were to do this job manually. But in a small fraction of time! So to anyone who has ever done this job manually in the past: just let PREQUAL do the job now!
The paper in Bioinformatics can be found here.
The software is freely available in GitHub.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in