Biomaterials for gene editing – facilitating CRISPR from the lab to the clinic
Published in Bioengineering & Biotechnology, Genetics & Genomics, and General & Internal Medicine

Since the first chimeric antigen receptor T cell therapy [Mullard 2017], now better known as CAR-T therapy was introduced in the form of a commercial product known as tisagenlecleucel to treat acute lymphoblastic leukemia; the clinical potential of genetic engineering for immunotherapy has rapidly risen. This method of precision medicine depends on the delivery of genes to modify T cells via lentivirus transduction; a topic that I relatively recently briefly outlined as a ‘new niche of biomaterials’ relative to immuno-materials that can regulate the immune system, and thereby attenuate disease mechanisms through the targeted activation or suppression of an immune response for immunotherapy and immunodiagnostics applications.
This post redefines the niche relative to biomaterials and microfluidics (another long-time interest of mine) to efficiently deliver gene-editing machinery into cells to create next-generation gene-editing platforms. These platforms are suited for therapeutic intervention in cancer immunotherapy and cardiac tissue engineering, with highlights of its impact for gene repair relevant to bottom-up engineer disease remodeling pathways in organoids.
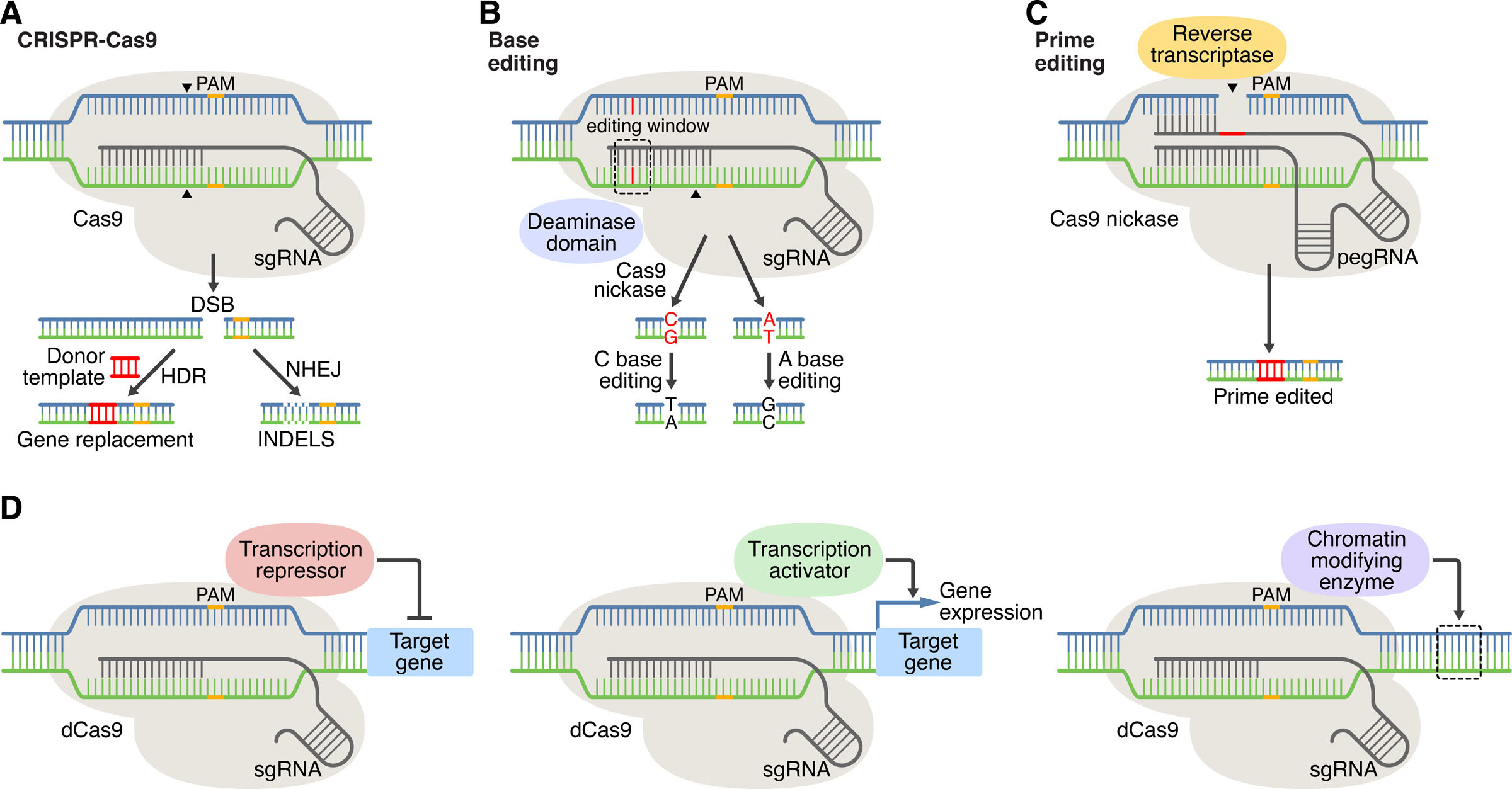
Figure 1: CRISPR-Cas systems for genome editing and beyond. A) In CRISPR editing, target recognition is mediated by base pairing of DNA with the sgRNA. Cas9 (CRISPR-associated protein 9) recognizes the PAM (protospacer adjacent motif) and generates DNA double-stranded breaks (DSBs). DSBs are repaired by nonhomologous end joining (NHEJ) or by homology-directed repair (HDR) in the presence of a donor template. B) base editing method, C) prime editing method, D) CRISPR-Cas mediated gene regulation and epigenetic modification. Credit: [Liu 2022].
A brief recap of the advent of CRISPR
The mechanism of clustered regularly interspaced short palindromic repeats is no stranger to the life sciences; highly popular in the past decade and broadly incorporated across the interdisciplinary fields of bioengineering, molecular biology, biochemistry, and biophysics. I too regularly covered this topic for years from CRISPRi or CRISPR-interference, to CRISPR-Cas9 and CRISPR Cas12a to highlight its versatility and advancements through the years.
Since its serendipitous discovery as a prokaryotic defense mechanism to cleave viral DNA [Ishino 1987][Jinek 2012], and its brilliant repurposing for eukaryotic cells [Zhang 2013],[Qi 2013], which eventually led to advanced DNA editing methods, including its FDA approved clinical applications – the CRISPR method has had a profoundly prolific ascent in the sciences; in what seems a relatively short timeframe [Mullard 2017]. The ubiquity of the method in the interdisciplinary sciences is even more so highlighted by perhaps an example of the quickest Nobel prize being awarded to the discovery in 2020, after its advent and emergence to the field predominantly in 2012 - to the very deserving scientist duo Emmanuel Charpentier, and Jennifer Doudna [Jinek 2012].
Encapsulating gene edited cell therapy within biomaterials and proof-of-concept CAR T-cell engineering.
Using biomaterials to deliver CRISPR-Cas9 for gene therapy holds immense potential to treat infectious diseases in an innovative approach that combines the advantages of gene therapy with protective biomaterials encapsulation [Dubey 2023]. Materials scientists can design biomaterials and particle properties to attain the desired non-viral nanocarrier features that can undergo degradation or hydrolysis hours after gene delivery. Such nanomaterials are composed of lipids or polymers to facilitate rapid biodegradation and release the intracellular biological cargo, while preventing potential cytotoxicities [Shannon 2022].
Biomaterial-mediated gene editing is suited for cancer immunotherapy, where loaded delivery carriers can travel through systemic circulation via active cellular uptake mechanisms for intracellular nucleic acid release with minimal cytotoxicity. Biomaterials for gene delivery to immune cells can be broadly classified as polymeric carriers, lipid-based carriers, and inorganic vesicles.
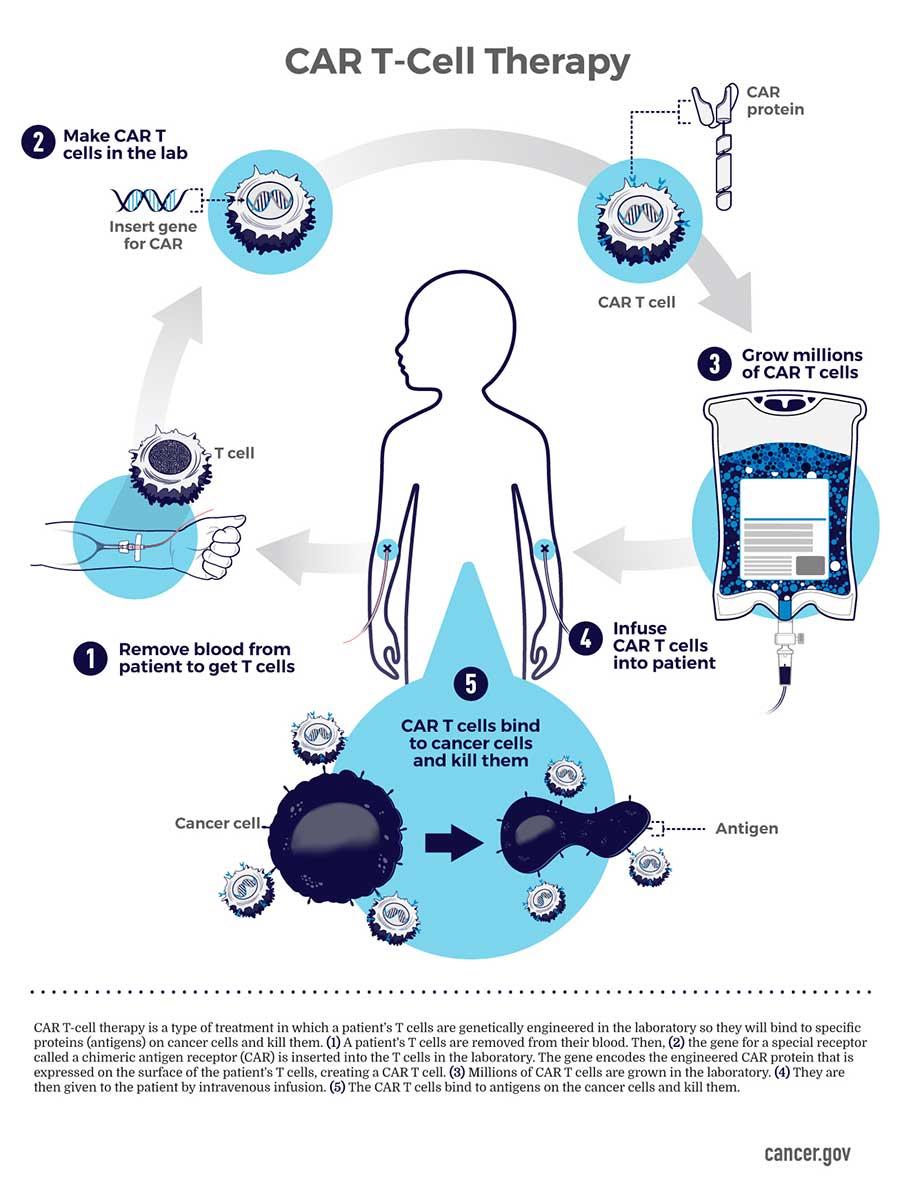
CRISPR is predominantly incorporated to engineer chimeric antigen receptor T-cells; traditionally isolated from a patient and engineered ex vivo to introduce a chimeric antigen receptor specific to the patient’s tumor cells, thereby expanded, and re-introduced to the patient to kill the tumor cell targets [Ellis 2021]. Bioengineers have developed cost-effective nano and microparticles, or implantable scaffolds for the local delivery of CAR-T cells to enhance their efficiency in solid tumors [Figure 2].
Chimeric antigen receptor T-cell therapies have revolutionized the field of cancer immunotherapy with genetically engineered tumor recognition, and the derivatives can effectively treat many other diseases [Niu 2023]. Additionally, metallo-supramolecules including metal-organic frameworks can self-assemble with the Cas9 nuclease and the guide RNA to deliver the CRISPR/Cas9 system for intracellular genome editing [Zheng 2021].
Microfluidics for gene editing
The next possibility of combining gene-editing with microfluidics is primarily focused on improving the efficiency of delivering gene-editing machinery into cells [Ahmadi 2020]. Microfluidics can generate multiple droplets at a very small scale to manipulate cells at a single level to facilitate the delivery of gene-editing machinery into cells. For example, bioengineers Hugo Singha and colleagues have developed an automated gene-editing platform with digital microfluidics and have knocked out the stable expression of a reporter gene in a small lung carcinoma cell line as proof-of-concept [Sinha 2018]. They then targeted an oncogene in the same cell line with a standardizing fluorescent image-based analysis method.
Another group similarly developed ‘cell squeezing’ – a device designed with microchannels to deform cells passing through the channels for transient membrane disruptions and for the diffusive intracellular uptake of cellular material [Sharei 2013]. When they compared the process with cell electroporation for intracellular delivery of genetic materials, the method of cell squeezing retained genetic expression profiles of the cells, much like the cohort of untreated cells to show mechanical-stress-preserved cell functionality for effective cell-based gene therapies [Shih 2015]. The range of gene editing methods via microfluidics are listed on table 1.
|
Type of device |
Purpose |
Strength |
Application |
|
|
Delivery of Cas9 and sgRNAs |
Channel microfluidics |
Plasmid and exogenous material delivery into the cells |
High cell viability and high delivery efficiency of various macromolecules into different cell types |
Shown for “hard-to-transfect” lymphoma cells and embryonic stem cells |
|
Digital microfluidics |
Automating gene-editing using digital microfluidics |
Fully automated pipeline of CRISPR knockout on chip (i.e., culture, edit, image-based analysis) |
Knocking out a stably expressing reporter and a RAF1 oncogene in a small lung carcinoma cell line |
|
|
Droplet-based microfluidics |
A lipoplex (cationic lipid-nucleic acid complex)-mediated single-cell transfection in a droplet-based microfluidic |
Significant increase of gene delivery efficiency via single-cell transfection going from ∼5% to ∼50% in K562, THP-1, and Jurkat cells |
Significant improvement in transfection efficiency for three suspension cell lines, i.e., K562, THP-1, and Jurkat |
|
|
Validation and expansion of the gene-edited cells |
Optical electrowetting |
Light-activated cell identification and sorting system |
3500 “NanoPens” for selecting and expansion of mammalian cells |
Single-cell manipulation, clonal expansion, and phenotypic analysis for primary T-cells in nanoliter volumes |
|
Hybrid microfluidics (i.e., combination of droplet and digital microfluidics) |
Selection, isolation, and expansion of successfully edited mammalian clones using a hybrid microfluidic system |
Individual addressability of single cells; in situ encapsulation of single cells in droplets for direct expansion |
On-demand selection, isolation, and expansion of gene-edited small lung carcinoma cells |
|
|
Potential for gene-editing |
Channel microfluidics |
Cell squeezing for the diffusive intracellular uptake of exogenous material |
Non-traditional means of transfection—i.e., cell squeezing—minimally invasive |
For the delivery of transcription factors toward cell-based therapeutics |
|
Digital microfluidics |
Electroporation on a digital microfluidic platform |
High on-chip transformation efficiency (8.6 ± 1.0 × 108 cfu μg−1) |
Electroporation for bacterial transformation |
|
|
Robotics |
Traceable single-cell isolation and detecting single cells using impedance profiling |
An in situ-based method for detecting the presence of single cells without fluorescence labeling |
A pipetting robot for single-cell isolation and detection |
|
|
Channel microfluidics |
Versatile intercellular delivery platform for hard-to-transfect primary stem and immune cells |
High delivery efficiency (up to 98%), user-friendly (single step operation), and high scalability (1 × 106 cells/min) |
Universal intracellular transfection platform for highly efficient transport of different biomolecules into hard-to-transfect primary cells |
Table 1: Microfluidic studies involved in or potentially used for gene-editing [Ahmadi 2020].
CRISPR for cardiac tissue engineering to treat heart disease
The RNA-guided CRISPR-Cas system [Knott 2018] can simplify genome editing to improve the accessibility of the method within tissue microenvironments for strategies of precision medicine. The discovery and applications of the CRISPR-Cas system can evolve beyond archaea and bacteria that bind and cleave foreign nuclei acids as an adaptive immune system [Liu 2022].
This has led to its adaptation to remodel mutations in mice and within human induced pluripotent stem cells. In cardiac engineering for instance, CRISPR-Cas9 has replaced embryonic stem-cell based homologous recombination as a routine tool to generate mutant mice with knock-out genes of interest, and is instrumental to identify complex genetic causes underlying cardiomyopathy, by modeling somatic gene mutations in the heart [Gifford 2019].
Aside from correcting damaging missense mutations, it is possible to conduct base editing to restore gene expression to maintain therapeutic gene editing. In order to deliver these genome editing components, the strategies must be safe, efficient, and made possible via nanoparticles and virus-like particles that enter the muscle and cardiovascular system to precisely attenuate the genetic basis of cardiomyopathies [Liu 2022] as a guide to genetic engineering in the future [Figure 3].
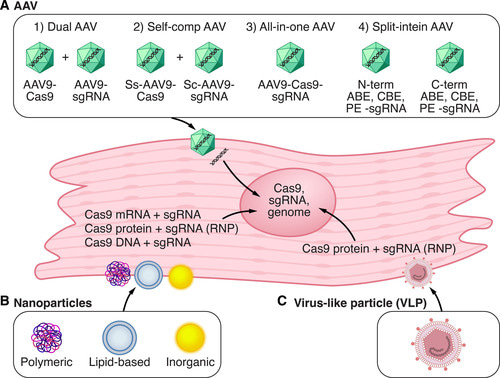
Figure 3: Strategies for in vivo delivery of CRISPR-Cas9 components in cardiac tissue: A) adeno-associated virus (AAV) is a common delivery vector for CRISPR-Cas9. B) nanoparticles can deliver Cas9 DNA, mRNA, or protein with single guide RNA into cells. C) Virus-like particles can deliver Cas9 components. ABE – adenine base editor, PE – prime editor. Credit: [Liu 2022].
Bottom-up engineering a disease remodeling pathway with CRISPR in organoids for gene repair.
Organoids can bridge the gap between 2-D cell lines and in vivo studies due to their 3-D organization and cellular heterogeneity to bring genome engineering a step closer to patients. The platform is well suited to effectively observe the construction and progression of isogeneic disease models to accurately qualify the impact of allelic disease variants in patients [Geurts 2023]. For instance, CRISPR-Cas9 mediated gene engineering acts by introducing double-stranded DNA breaks into the genome for gene knockout studies for the precisely targeted introduction of exogenous DNA.
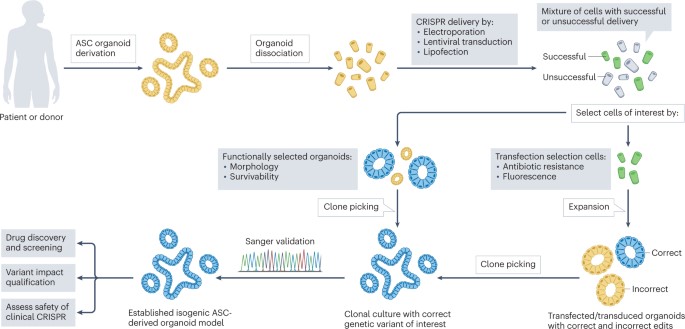
This provides a good model system to study human health and disease. The genome engineering method provides an isogenic model in a biological microenvironment to bottom-up engineer and investigate the cause, onset, and treatment of human diseases in the lab simply by using adult-stem-cell-derived organoids suited for clinical translation [Figure 4]. Such ex vivo repaired organoids can also be transplanted back into patients to relieve the disease phenotypes.
Researchers have studied six classes of Cas genes, of which the class II CRISPR system that includes the Cas9 variant is the most significant. By hijacking the endogenous DNA repair pathways, CRISPR can be incorporated for genome engineering in a single-guide RNA-mediated method [Jinek 2012]. Bioengineers can conduct multiple CRISPR-screens in a single experiment, for example, researchers have performed a small targeted CRISPR screen in human intestinal organoids to map the genetic dependencies during colorectal cancer progression [Geurts 2023]. To accurately model mutations observed in cancer patients, researchers must aim beyond simple CRISPR-Cas9 mediated knockouts via indel formation. For instance, different mutations in the same cancer gene can have significantly different effects as seen with TP53 gene mutations in myeloid malignancies [Boettcher 2019] and the KRAS codon in isogenic cell lines [Stolze 2014] to highlight the necessity of DNA double-strand break-free genome engineering in organoids.
Delivering gene editing to repair hereditary diseases.
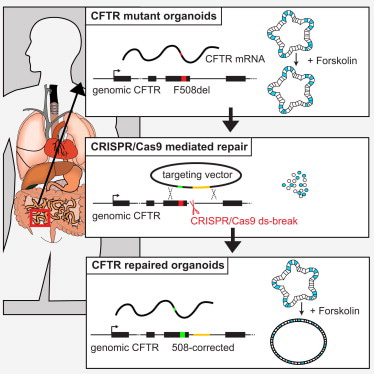
I wrap-up this article by describing disease modeling beyond cancer, in the lab, and by highlighting the capacity to treat hereditary diseases via gene editing, for the first time, in the clinic [Geurts 2023]. CRISPR-mediated gene repair has new-found promise due to its clinical potential relative to the transplantation of gene-modified organoids back to patients to complement whole-organ transplantation [Hu 2018]. On the same vein, cystic fibrosis is the first known hereditary disease that can be repaired in human stem cells by using CRISPR-Cas9 mediated genome engineering.
The disease results from a variety of mutations that occur in the cystic fibrosis transmembrane conductance regulator gene, which commonly includes the deletion of phenylalanine-508 [Sosnay 2013] that can be repaired via prime-editing [Shwank 2013] [Figure 5]. Life scientists have also shown the capacity for prime-editing in patient-derived intestinal organoids to restore the most common mutations in intestinal organoids. Prime editing is a preferred strategy since the reverse-transcriptase template can be designed to exclusively include the edit of interest [Geurts 2023].
The process of delivering genome-editing agents into organoids is a more complex task than with 2-D cell lines. While lentiviral transduction can be an efficient strategy when compared to simple lipofection, functional selectivity is another possibility based on the morphology or the swelling response of intestinal organoids that carry a wild-type, or mutant gene [Geurts 2023]. In the absence of functional selection, transfection selection is another preferred strategy through flow-assisted cell sorting based on fluorescence or through co-transfection. It is also possible to deliver CRISPR-Cas components via adeno-associated viruses, while nanoparticles can deliver Cas9 DNA, mRNA, or proteins with single guide RNA into cells [Liu 2022] [Figure 6].
Outlook – the bigger picture
As always, fundamental details form the bigger picture and is no exception in this instance to understand the profound influence of gene editing advances from the lab-to-the clinic, for the possibility of treating cancer, and to repair hereditary diseases based on FDA approval, in a short span of time. Since the advent of the CRISPR-Cas9 gene editing method and since repurposing it from an inherent prokaryotic defense mechanism to eukaryotic cell lines, the technique has thus far made significant strides in scientific discovery.
The overarching theme of this article is to examine the efficiency of delivering gene edited organoids and engineered single cell lines through microfluidics-based encapsulation or via biomaterial-assisted methods to treat patients. Efficient biomaterials or nanomaterials undergo degradation upon gene delivery to prevent cytotoxicities as intracellular biological cargo during the repair of hereditary diseases such as cystic fibrosis, sickle cell disease, beta thalassemia, and even leukemia [Sheridan 2023]. With revolutionary impacts of gene editing brought to light to correct cardiovascular disease and the advent of chimeric antigen receptor T-cell therapies; predominantly in the field of cancer immunotherapy as discussed herein.
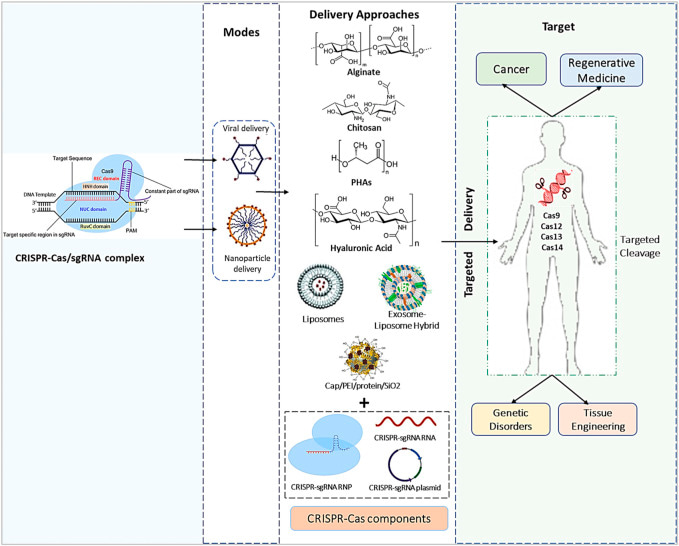
Figure 6: The bigger picture of the CRISPR/Cas9 payload’s genetic engineering processes, effective diagnostics methods, and in vivo delivery techniques used in numerous types of medical treatment and medication delivery systems. [Dubey 2023]
The rapid advances of gene editing methods and their convergence with effective gene delivery has allowed scientists to correct disease-causing genetic errors in ways that were hitherto only imagined. While technical, medical, and ethical challenges remain to resourcefully harness the power and potential of the technology to treat a myriad of diseases by correcting disease causing genetic errors at large, the biomedical future of this method has a very promising outlook.
Header Image is a variant of Crispr upgrade can make genome editing better and safer via the New Scientist
References
- Mullard A. et al. FDA approves first CAR T therapy, Nature Reviews Drug Discovery, doi: https://doi.org/10.1038/nrd.2017.196
- Liu N. et al. CRISPR Modeling and Correction of Cardiovascular Disease, Circulation Research, doi: https://doi.org/10.1161/CIRCRESAHA.122.320496
- Ishino Y. et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product, Journal of Bacteriology, doi: 10.1128/jb.169.12.5429-5433.1987
- Jinek M. et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity, Science Advances, doi: 1126/science.1225829
- Ran F. et al. Genome engineering using the CRISPR-Cas9 system. Nature Protocol, doi: https://doi.org/10.1038/nprot.2013.143
- Qi L. et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression, Cell, doi: 1016/j.cell.2013.02.022
- Dubey A. et al. Biomaterials-mediated CRISPR/Cas9 delivery: recent challenges and opportunities in gene therapy, Frontiers in Chemistry, doi: 10.3389/fchem.2023.1259435
- Shannon S. et al. Approaches towards biomaterial-mediated gene editing for cancer immunotherapy, Biomaterials Science, doi: https://doi.org/10.1039/D2BM00806H
- Ellis G. et al. Genetic engineering of T cells for immunotherapy, Nature Reviews, Genetics, doi: 10.1038/s41576-021-00329-9
- Niu H. et al. Biomaterials for chimeric antigen receptor T cell engineering, Acta Biomaterialia, doi: https://doi.org/10.1016/j.actbio.2023.04.043
- Zheng Q. et al. Nanoscale metal–organic frameworks for the intracellular delivery of CRISPR/Cas9 genome editing machinery, Biomaterials Science, doi: https://doi.org/10.1039/D1BM00790D
- Ahmadi F. et al. Is microfluidics the “assembly line” for CRISPR-Cas9 gene-editing? Biomicrofluidics, doi: 10.1063/5.0029846
- Sinha H. et al. An automated microfluidic gene-editing platform for deciphering cancer genes, Lab Chip, doi: 10.1039/c8lc00470f
- Shih S. et al. A Versatile Microfluidic Device for Automating Synthetic Biology, ACS Synthetic Biology, doi: 10.1021/acssynbio.5b00062
- Knott G. et al. CRISPR-Cas guides the future of genetic engineering, doi: 10.1126/science.aat5011
- Gifford C. et al. Oligogenic inheritance of a human heart disease involving a genetic modifier, doi: 10.1126/science.aat5056
- Geurts M. et al. CRISPR engineering in organoids for gene repair and disease modelling, Nature Reviews Bioengineering, doi: https://doi.org/10.1038/s44222-022-00013-5
- Boettcher S. et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies, doi: 10.1126/science.aax3649
- Stolze B. et al. Comparative analysis of KRAS codon 12, 13, 18, 61 and 117 mutations using human MCF10A isogenic cell lines, Scientific Reports, doi: https://doi.org/10.1038/srep08535
- Hu H. et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids, Cell, doi: https://doi.org/10.1016/j.cell.2018.11.013
- Sosnay P. et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene, Nature Genetics, doi: https://doi.org/10.1038/ng.2745
- Shwank G. et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients, Cell Stem Cell, doi: https://doi.org/10.1016/j.stem.2013.11.002
- Sheridan C. et al. The world’s first CRISPR therapy is approved: who will receive it? Nature Biotechnology, doi: https://doi.org/10.1038/d41587-023-00016-6
Follow the Topic
Your space to connect: The Primary immunodeficiency disorders Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine, Immunology, and Diseases!
Continue reading announcement




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in