
The paper in Nature Ecology & Evolution is here: http://go.nature.com/2fNGlBY
"To be honest, I don't think it's a good idea, .... but yes, something could be found with that analysis. Please go forward with that analysis."
I have to confess that I was less than eager to agree with the analysis proposed by one of my collaborators, Song Guo. However, that analysis eventually provided an important hint for the main findings of our paper. (a hint for why vertebrates remained their mid-embryonic phase conserved during evolution).
General relationship between ontogeny and phylogeny
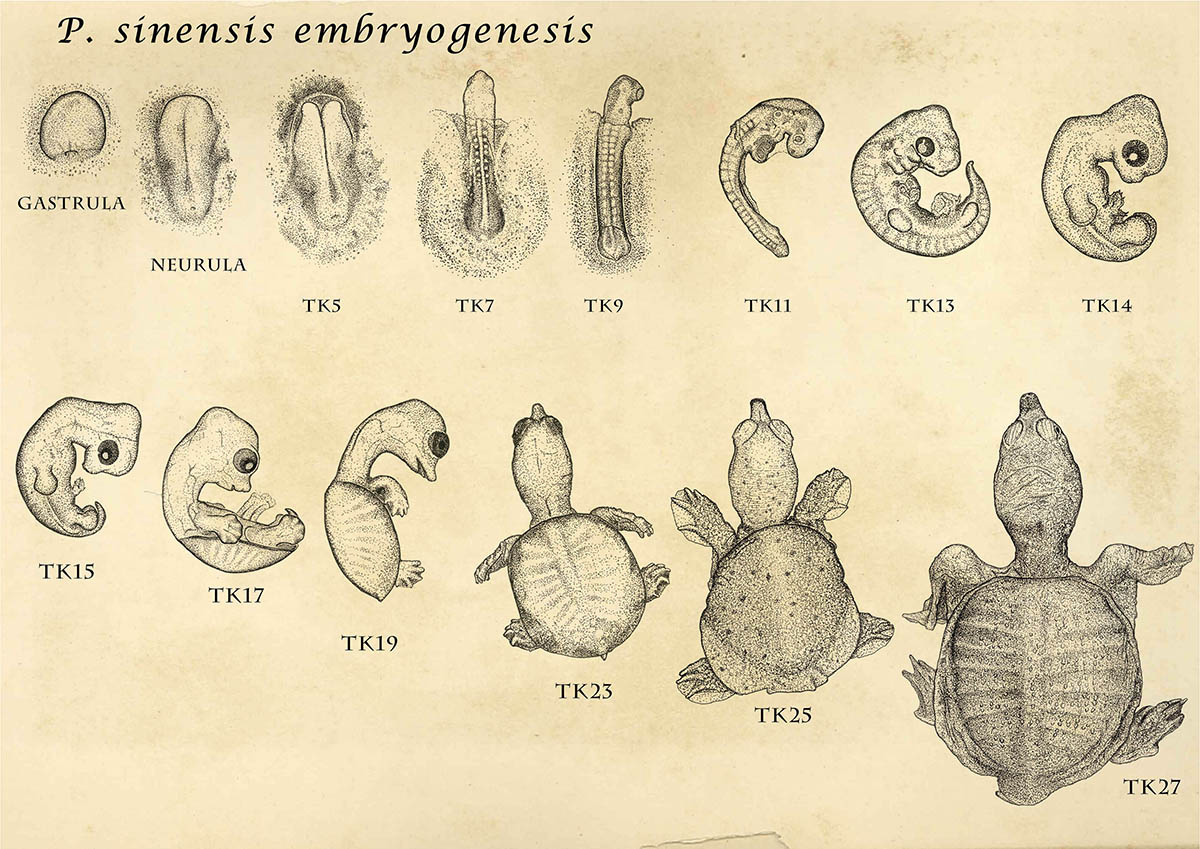
Recent comparative transcriptomic studies, including ours, supported somewhat counter-intuitive model for the general relationship between ontogeny (embryogenesis) and phylogeny (evolution), namely, the developmental hourglass model (first proposed by Dr. Denis Duboule). Considering that earlier embryonic processes establish information required for the later developmental events, it appear reasonable to assume that earlier stages are more difficult to change during evolution, and thus become more evolutionarily conserved than later stages. However, the developmental hourglass model predicted that it is mid-embryonic stages (especially stages that establish bodyplan of each animal phylum) that are conserved the most. One of the main goals of our study was to find whether the species in the same phylum would really fit to this model (as has been predicted), and we have selected and studies 8 chordate species for this purpose.
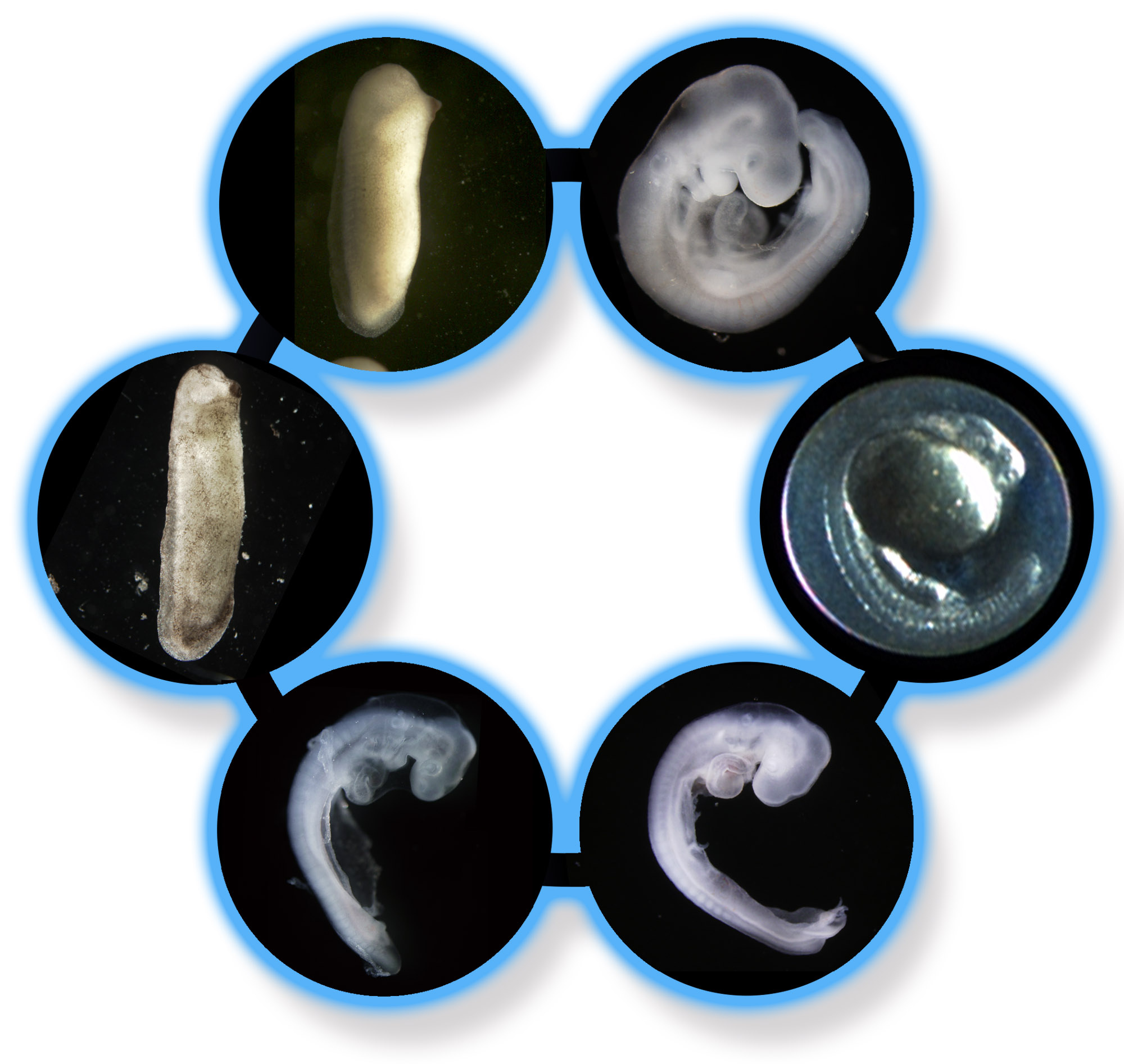
In the meanwhile, the idea proposed by Song Guo and colleagues in Philipp Khaitovich's lab was to find genes specifically expressed at the conserved stages in vertebrates to find a hint for a "potential mechanism" that made embryos to follow the hourglass model. However, I have to say that was reluctant to go with this idea. Actually, I had this kind of suggestions many times at meetings. The reason why I avoided performing this analysis is simply because I thought it was asking too much.
The data we had is gene expression profiles obtained from early-to-late embryos of 8 chordate extant species. Even though we found genes commonly and specifically expressed at the conserved stages, what can we say about the mechanism? It's just an 'outcome' of hundreds of millions of evolution, and deducing exact mechanism from the outcome of evolution (gene expression) would be just too much. Moreover, even though recent studies have reported similar expression profiles among different species and supported the hourglass model, we still do not know what that really means. One expectation is that the similar gene expression profiles would be reflecting similar composition of homologous cells among different species, however, no one have yet tested this thoroughly. In this situation, I thought going into evolutionary mechanism with gene expression data would be a bit over-eager. However, it was this assumption that turned out to be too much.
Song soon reported us that she did not find anything expected. Despite the fact that the conserved organogenesis stages are phase of active morphogenesis and many genes should be working, there was much less stage-specific genes. "Your were right, Naoki." she continued. However, this sounded very strange, as we already had basic results that confirmed numbers of genes expressed during development do not differ largely (increases almost constantly without showing any major drop at mid-embryonic stages). With this "less stage-specific genes at mid-embryonic phase", we soon realized that genes expressed at the conserved stages should also be expressed at other developmental stages. As we reported in our paper, genes expressed at mid-embryonic stages were actually expressed at variety of developmental stages and tissues. Moreover, genes expressed in many stages were predicted to have more predicted transcription factor binding sites, more protein-protein interactions, and tended to be essential for normal embryogenesis. What is important, developmental stages that had higher ratio of the pleiotropically expressed genes tended to be evolutionarily conserved.
One concept that could reasonably explain these results were "pleiotropic constraint", and we thus concluded as pleiotropic constraint could be contributing to restrict the diversification of mid-embryonic stages. If this was true, it would also mean that reusage of genes would be a double-edged sword for evolutionary diversification. While gene recruitment (called co-option) is known to have contributed to evolve novel traits, it may also mean that gene recruitment constrains the evolutionary changes of traits that utilizes the same genetic machinery. Does it mean that more complex (at regulatory level) organism have less evolvability? As we discussed in detail in the manuscript, there are many questions to be answered regarding these ideas. We would also have to do more than analyzing embryos, even though they harbor significant amount of hints for how they have evolved. We could, or I would say should, still be oversimplifying the nature as I actually did in the early phase of this project.
The paper in Nature Ecology & Evolution is here: https://goo.gl/e9XqZY





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in