Decoding Corn Yield Efficiency: How Deep Neural Networks Can Help Build a More Sustainable Future
Published in Earth & Environment, Ecology & Evolution, and Microbiology
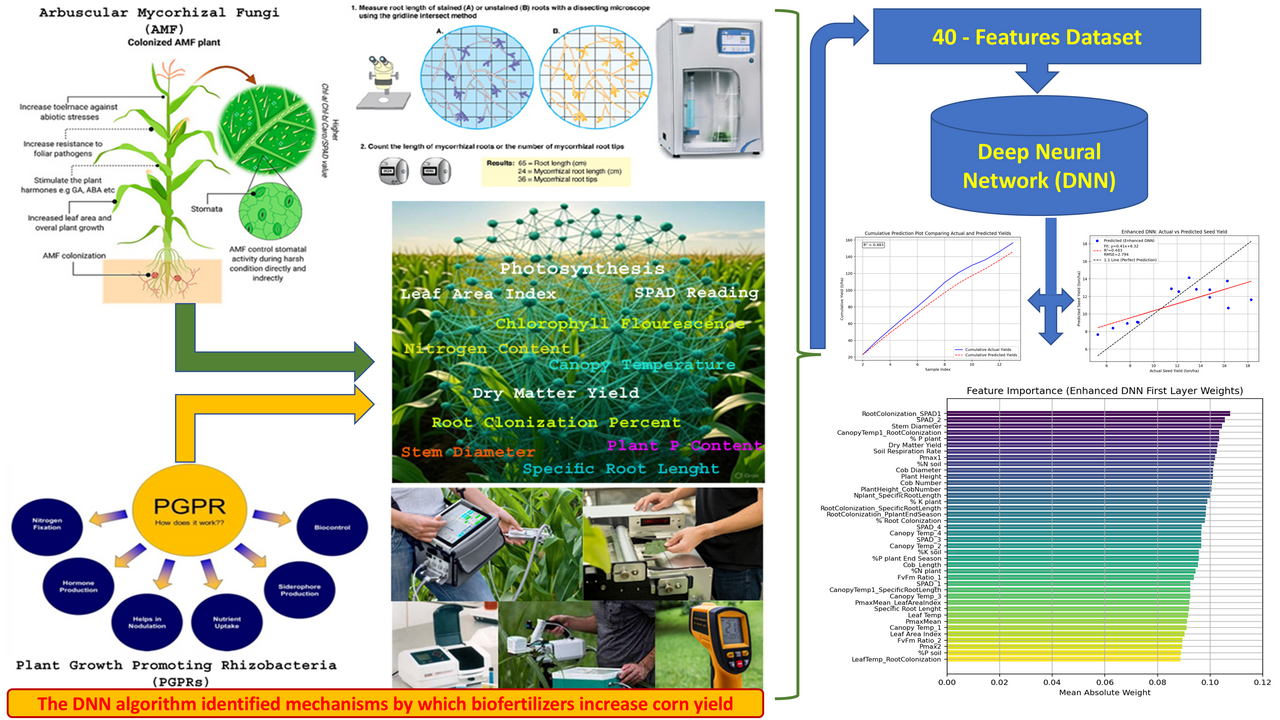
The degradation of fundamental natural resources, the decline in biodiversity, the ever-growing population, and the rise of cultural and geopolitical conflicts have collectively rendered our planet’s environment more unstable than ever before. For the first time in human history, our shared ecosystem faces the tangible risk of total collapse. In this context, it has become imperative to reduce the environmental costs of food production while simultaneously ensuring that the growing global population can access food in safer and healthier ways. One of the most essential pathways toward this goal lies in improving resource-use efficiency on farms. By enhancing how plants absorb and utilize water, nutrients, and energy—and by understanding the eco-physiological pathways through which these resources are converted into food—we can minimize input usage while ensuring the sustainable exploitation of soil, water, and plant resources.
In this study, titled “Enhanced Deep Neural Network with Interaction Features for Corn Seed Yield Prediction: Uncovering AgroEcophysiological Relationships,” published in Discover Agriculture (Springer Nature), we sought to bridge the gap between ecophysiology and artificial intelligence to improve the sustainability of agricultural systems. Our motivation was rooted in a simple yet powerful question: Can we teach machines not only to predict yield but also to understand the biological logic behind it?
The Motivation Behind the Work
Modern agriculture generates enormous volumes of data—from soil nutrients and microbial activity to canopy temperature and rainfall patterns. However, these data streams often remain fragmented and underutilized. Conventional statistical models struggle to capture the complex, nonlinear interactions between biological and environmental processes that determine crop yield. At the same time, purely black-box AI models, while powerful, often lack interpretability, making them difficult to trust or apply in real-world agronomic decision-making.
This tension between complexity and interpretability became the foundation of our research. We aimed to create a deep learning framework capable of learning hidden patterns among agronomic variables while also providing explainable insights into the ecophysiological mechanisms underlying crop productivity.
Our Approach: From Data to Discovery
We developed an Enhanced Deep Neural Network (EDNN) architecture that integrates interaction features—mathematical representations of how two or more environmental or physiological variables jointly influence yield outcomes. These interaction features allow the network to model cross-effects such as:
-
how soil phosphorus availability interacts with canopy temperature,
-
how microbial colonization modifies nutrient uptake efficiency,
-
and how rainfall and solar radiation jointly affect kernel formation.
By incorporating such features, our EDNN acts not just as a prediction tool but as a digital reflection of the plant’s biological logic.
The model was trained and validated on large-scale experimental data from corn fields subjected to different biofertilizer treatments (including arbuscular mycorrhizal fungi and plant growth-promoting rhizobacteria). These treatments are known to influence resource-use efficiency and root–soil interactions—a critical component of sustainable agriculture.
Overcoming Challenges
Developing this framework was far from straightforward.
One major challenge was data heterogeneity—each dataset came from experiments under different climatic and soil conditions. Normalizing and harmonizing these variables without losing their biological meaning required both agronomic expertise and computational precision.
Another difficulty was avoiding overfitting while preserving model interpretability. To address this, we incorporated dropout layers, early stopping, and SHAP (SHapley Additive exPlanations) analysis to ensure that the model’s predictions were biologically sound and explainable.
But perhaps the greatest challenge was conceptual: finding a way to speak both languages—the language of ecology and the language of algorithms—and allowing them to meet in a space where both make sense.
Key Findings and Insights
The EDNN achieved a substantial improvement in yield prediction accuracy compared to traditional machine learning models. More importantly, its explainability revealed novel agroecophysiological relationships that had not been previously quantified.
For example, the model uncovered that under biofertilizer application, soil respiration and canopy temperature exhibit a compensatory dynamic—when microbial activity enhances nutrient mineralization, plants require less transpirational cooling to maintain optimal photosynthetic performance. Such insights highlight the bio-efficiency mechanisms through which natural microbial associations can replace or complement chemical inputs.
This fusion of AI interpretability and ecophysiological reasoning offers a pathway toward “intelligent sustainability”—a future where technology helps us understand nature’s own logic rather than override it.
Why It Matters
Global agriculture stands at a crossroads. The challenge of feeding nearly 10 billion people by 2050 cannot be met with incremental improvements alone. What we need are transformative frameworks that connect disciplines—bringing together biology, data science, and environmental ethics.
Our research contributes a step in that direction. It shows that artificial intelligence, when guided by ecological principles, can become a tool for biological discovery rather than just automation. By learning from the plant’s inner logic, we can move toward a form of agriculture that is both productive and regenerative.
Looking Ahead
This work opens the door to a new generation of hybrid models—systems that combine deep learning, explainable AI, and ecophysiological simulation. The next step is to apply similar frameworks to multi-crop systems, assess regional adaptation strategies, and develop decision-support tools for farmers in arid and semi-arid regions.
Ultimately, this study is not just about algorithms or datasets; it’s about a vision. A vision where data-driven intelligence becomes an ally of ecological wisdom—helping humanity cultivate food systems that sustain both people and the planet.
Follow the Topic
-
Discover Agriculture

This journal covers basic science and applied research relating to the agricultural sector in areas such as crop and soil sciences, environmental science, and advances in relevant technologies.
Related Collections
With Collections, you can get published faster and increase your visibility.
Resilient Food Systems: Underutilized Crops, Improved Agricultural Practices, and Policy Levers for Food Security
The FAO's The State of Food Security and Nutrition in the World (SOFI) 2025 report indicates that approximately 2.3 billion people worldwide experienced moderate or severe food insecurity in 2024 – an increase of 335 million since 2019 (nearly 28% of the global population). This escalating crisis demands resilient and diversified food systems, as traditional agriculture faces pressures from climate change, land degradation, and volatile markets, disproportionately impacting vulnerable populations.
This research area focuses on enhancing food security by strategically integrating underutilized crops (teff, fonio, bambara groundnut, etc.), improved agricultural practices, and supportive policies. Underutilized crops offer a significant, untapped resource for diversifying food production and improving nutrition, as they are often nutrient-rich and well-adapted to local environments. Sustainable practices like agroforestry and climate-smart agriculture are crucial for resource efficiency and environmental resilience.
However, these advancements are insufficient without effective policy levers, particularly regarding land tenure security and market access. Policies supporting agricultural extension and R&D are also vital. This Collection aims to identify pathways to resilient, equitable, and sustainable food systems by exploring the interplay between underutilized crops, improved agricultural practices, and policy interventions.
The Collection addresses food insecurity exacerbated by climate change, biodiversity loss, and staple crop reliance. We explore solutions through: underutilized crops as climate-resilient alternatives; improved practices like agroecology for sustainable productivity; and policy levers to promote adoption. By integrating these, we can build resilient and diverse food systems ensuring food security and environmental sustainability.
This Collection explores pathways toward resilient food systems through underutilized crops, innovative agricultural practices, and effective policy frameworks. This Collection invite contributions addressing:
• Nutritional Composition & Bioaccessibility: Analyzing macro-and micronutrient content, bioavailability, and processing impacts on nutritional value in underutilized crops.
• Genetic Improvement & Breeding: Developing improved, climate-resilient varieties with enhanced yield and nutrition through breeding and genetic analysis.
• Optimizing Agronomic Practices: Enhancing productivity and sustainability in underutilized crop cultivation via efficient irrigation, soil management, nutrient use, and pest/disease control.
• Climate-Resilient Farming & Diversification: Evaluating climate-smart practices and diversified systems using underutilized crops to mitigate climate change impacts.
• Postharvest Handling & Value Addition: Investigating innovative technologies to reduce food loss, extend shelf life, and create value-added products from underutilized crops.
• Market Development & Consumer Acceptance: Researching consumer preferences and effective strategies to integrate underutilized crops into diverse diets and markets.
• Policy & Regulatory Frameworks: Analyzing policies (subsidies, incentives, labeling) to support the adoption, production, and commercialization of underutilized crops.
• Land Governance & Resource Rights: Examining land tenure security, resource rights, and equitable access for smallholders involved in underutilized crop cultivation.
• Socioeconomic Impacts: Assessing impacts on income, livelihoods, food security, and nutrition for vulnerable populations, with attention to gender and inclusion.
• Traditional Knowledge & Modern Science: Integrating traditional knowledge with modern science for sustainable and resilient systems using underutilized crops.
Keywords: Food Security, Resilient Food Systems, Sustainable Agriculture, Agricultural Policy, Climate Change Adaptation, Underutilized Crops, Livelihoods, Rural Development, indigenous crops
This Collection supports and amplifies research related to SDG 1, SDG 2, SDG 12, and SDG 13.
Publishing Model: Open Access
Deadline: Jul 31, 2026
Resilient Rainfed Smallholder Farming Systems for Sustainable Development
Rainfed smallholder farming systems underpin food security, rural livelihoods, and ecological balance across much of sub-Saharan Africa and the Global South. Yet these systems remain highly vulnerable to climate variability, land degradation, declining soil fertility, and market volatility. Addressing these interconnected challenges requires innovative, context-specific, and systems-based solutions that increase productivity and enhance resilience, equity, and sustainability. This collection aims to gather cutting-edge research and transformative approaches that strengthen the ecological and socioeconomic foundations of rainfed agriculture. Submissions should explore the integration of sustainable land management, climate-smart innovations, and inclusive governance structures in rainfed smallholder contexts. Papers may cover a range of themes, including but not limited to: agroecological intensification strategies such as intercropping, mulching, or conservation tillage; soil and water conservation practices adapted to dryland environments; the use of digital agro-advisory tools and participatory extension models; farmer-led experimentation and co-creation of knowledge; gender and youth-responsive innovations; and enabling policy or institutional mechanisms that foster adoption and scalability. Contributions employing robust data, interdisciplinary perspectives, and clear pathways to real-world impact are especially encouraged.
Keywords: Agroecological intensification, Climate Resilience, Soil and water conservation, Low-input agriculture, Nature-based solutions, Digital agro-advisory services, Participatory innovation, Land degradation neutrality, Climate-smart agriculture
This Collection supports and promotes research aligned with SDG 1, SDG 2, SDG 5, SDG 13 and SDG 15.
Publishing Model: Open Access
Deadline: Mar 12, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in