Digging into the syntrophic origin of eukaryotes
Published in Microbiology

The incredible diversity of eukaryotes, from the unicellular algae and protozoa to the multicellular plants, fungi and animals, sparks fascination. How did this explosion of morphologies, structures and behaviors come to be? The more we dig into the murky origin of eukaryotes the more we realize that eukaryotes have well-anchored prokaryotic roots and that their evolutionary history started as a story of cooperation and metabolic trade-off in the oxygen-poor environments of the early Proterozoic, over 2 billion years ago.
By the late 1970s, molecular phylogenetic evidence made the scientific community embrace the symbiotic origin of mitochondria (from alphaproteobacteria) and chloroplasts (from cyanobacteria) so eagerly popularized by Lynn Margulis, but the symbiotic origin of the eukaryotic cell from archaea and bacteria took much longer to be accepted. At that time, Carl R. Woese showed that archaea, then a poorly known group of extreme environment-loving prokaryotes, formed an independent domain of life. The first rooted phylogenetic trees of the three domains of life (archaea, bacteria, eukaryotes) sustained the prevailing view that modern eukaryotes arose from an independent proto-eukaryotic lineage sister to archaea that evolved most eukaryotic features (including a complex cytoskeleton, endomembranes, nucleus and phagocytosis) before it engulfed the alphaproteobacterial ancestor of mitochondria. Later phylogenomic analyses with better methods and including more taxonomic sampling started to suggest that eukaryotes might derive from within archaea but it was the discovery of the Asgard archaea that definitely shifted mainstream views on eukaryogenesis towards models proposing an archaea-bacteria symbiosis at the origin of the eukaryotic cell. Why?
Asgard archaea share more and more similar genes with eukaryotes than other archaea and phylogenomic analyses of conserved genes place eukaryotes within this archaeal group. This implies that there were only two primary domains of life, archaea and bacteria, and that eukaryotes are mergers, a secondary domain derived from (at least) two ancestors, one Asgard archaeon and one alphaproteobacterium (future mitochondrion). At the same time, comparative analyses of their metagenome-assembled genomes suggest that most Asgard archaea (and also their ancestors) are versatile hydrogen producers or consumers and entertain syntrophy with hydrogen-scavenging microbes in the oxygen-poor environments where they mostly thrive. This was recently confirmed by the first cultivation of one Asgard archaeon, Prometheoarchaeum syntrophicum, which grows in syntrophy with either a sulfate-reducing deltaproteobacterium, a methanogenic archaeon or both.
This strongly revitalizes early models proposing a hydrogen-based syntrophy at the origin of eukaryotes. One of such models was the Syntrophy hypothesis (1998) which, based on solid microbial ecology grounds, postulated that eukaryotes derived from the endosymbiosis of a methanogenic archaeon within a fermentative deltaproteobacterium from which it scavenged hydrogen before a versatile aerobic and methanotrophic alphaproteobacterium joined the consortium. However, the discovery of Asgard archaea imposes new constraints on eukaryogenetic models. In this new context and based on well-known microbial ecological interactions in redox transition environments, we have updated the Syntrophy hypothesis (https://www.nature.com/article...), making the inter-partner metabolic exchange shift from a hydrogen-and-methane (HM) transfer towards a hydrogen-and sulfur-species (HS) transfer.
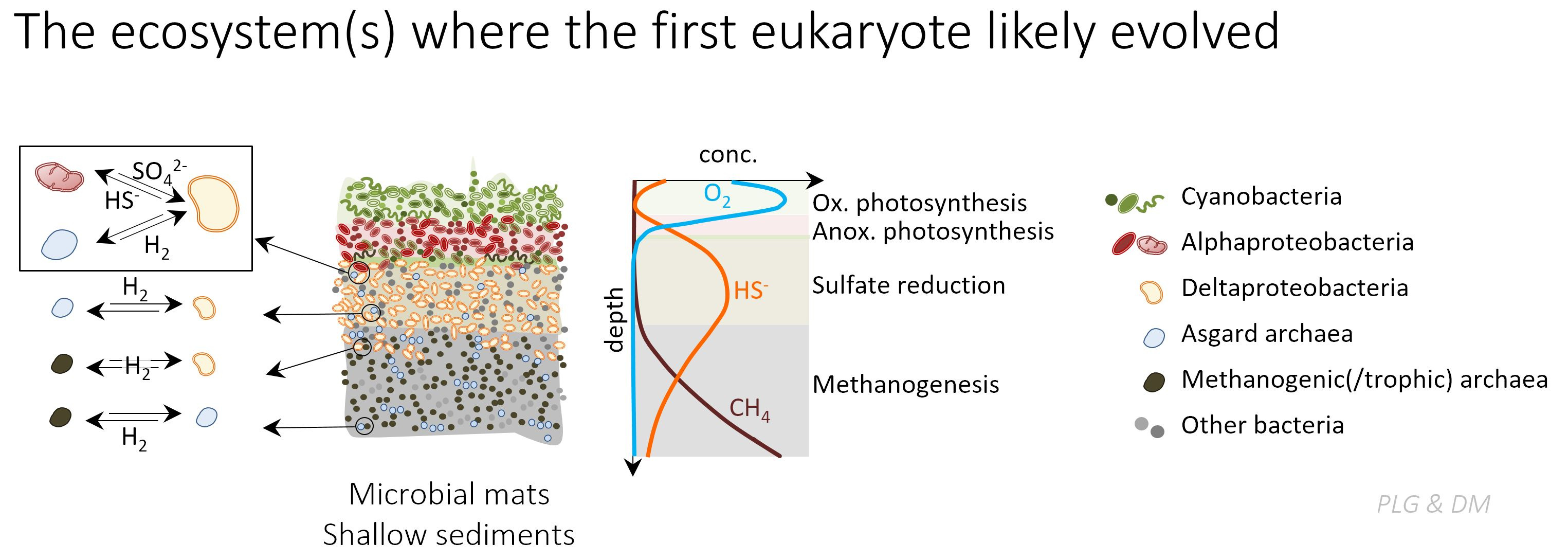

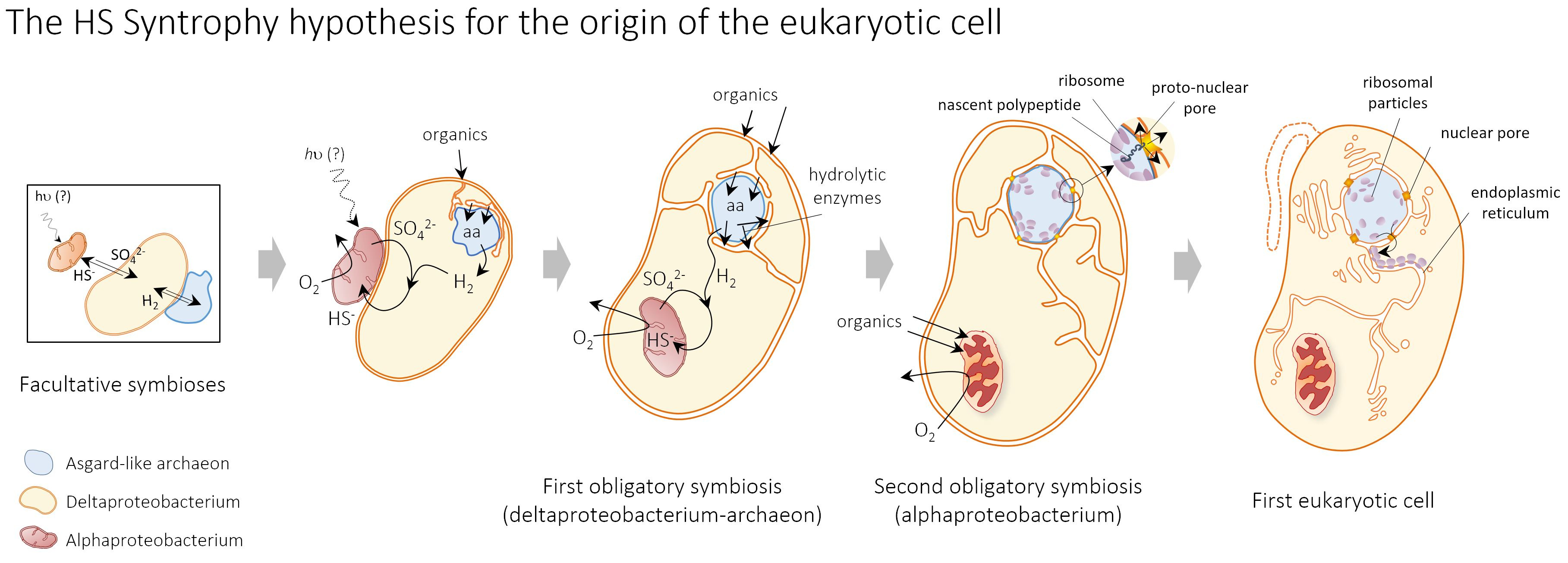
In the HS Syntrophy model, eukaryotes evolved in microbial mats or shallow sediments from the endosymbiosis of a hydrogen-producing Asgard archaeon within a hydrogen-scavenging sulfate-reducing deltaproteobacterium. A second endosymbiosis of a sulfide-oxidizing alphaproteobacterium that recycled sulfur-species in the consortium, and was potentially an anoxygenic photosynthesizer using sulfide as electron donor, evolved into the mitochondrion. The higher average complexity of the eukaryotic cell resulted from the physical integration of prokaryotic cells combined with extensive gene and genome shuffling. The HS Syntrophy model is one of the most comprehensive proposed so far as it accounts for the evolution of the endomembrane system, metabolism and genome. We compare it to other current symbiogenetic models and highlight differences and open questions for the future.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in