Does bottom trawling activity impact the benthic microbial biogeography?
Published in Earth & Environment and Ecology & Evolution

Bottom trawling is an industrial fishing practice by which heavy chains and nets are dragged over the seafloor. By landing approximately a quarter of all wild-caught seafood, bottom trawling provides access to a major food source. However, it also represents the most extensive anthropogenic disturbance that seabed habitats experience, as it physically reworks the upper sediment layers, re-suspends large amounts of sediment, injures and kills benthic macrofauna and alters biogeochemical processes and the sequestration of carbon.
The shallow shelf North Sea is one of the most heavily trawled regions on our planet. Given the high environmental impact of trawling, clever management and spatial planning of trawl fisheries in areas such as the North Sea is very important. For this reason, the German federal government is currently instating marine protected areas (MPAs) where the use of mobile bottom touching gears will be prohibited. Our research is part of Sustain Mare, an exciting joint project among several research institutes funded by the ministry of education and research (BMBF), which aims to monitor and study the changes in these MPAs and contribute in general to a better understanding of bottom trawling impacts.
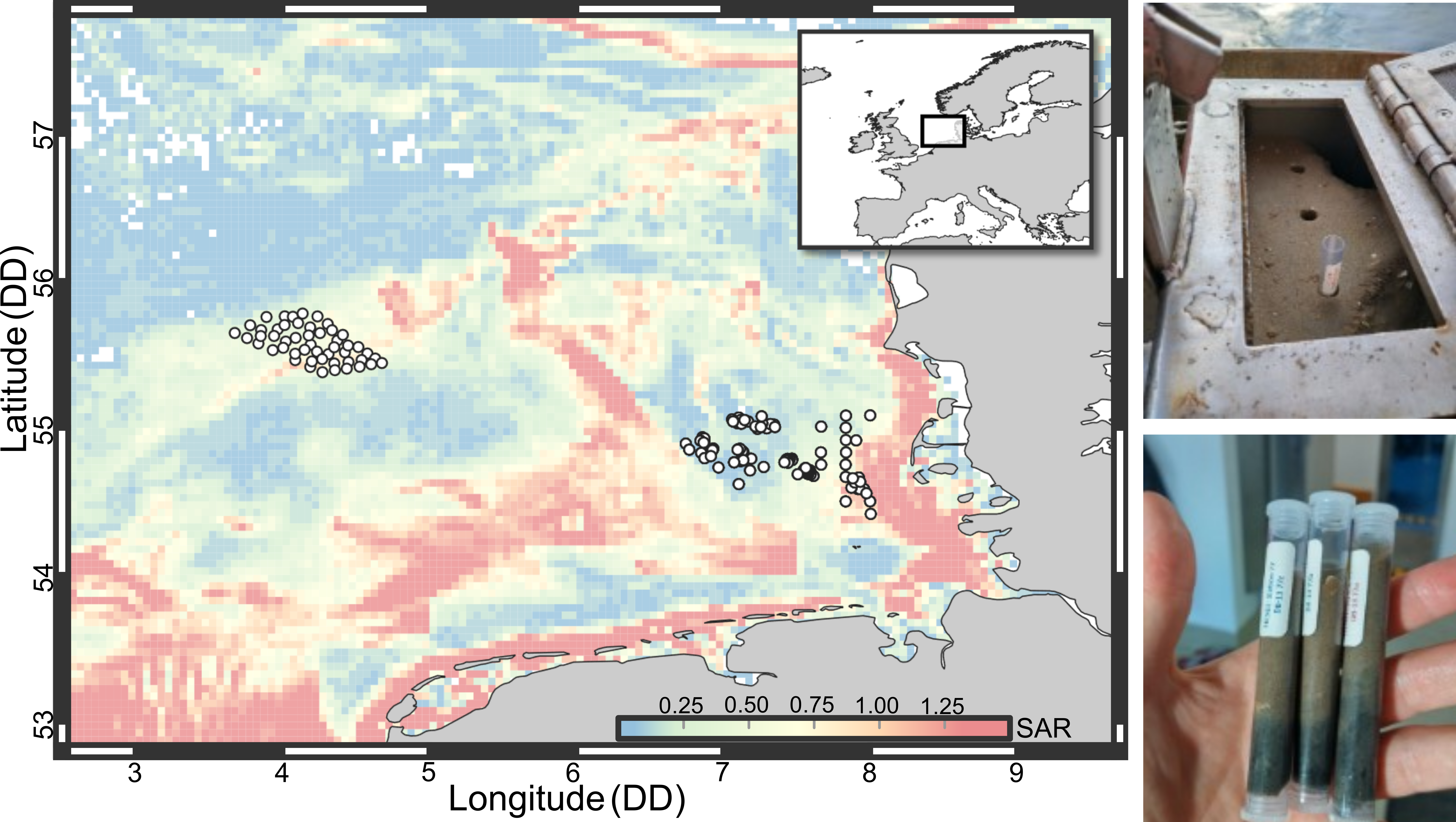
The average trawling intensity is expressed in swept area ration per year and displayed as a heatmap in the
background.
We are interested in the biogeography of marine microbes at larger scales. Microbes are small and occur in complex communities. But they are distributed across the planet. When we zoom out to scales of kilometers, hundreds of kilometers and beyond, what patterns arise in the microbial biogeography? And do human activities such as bottom trawling affect such larger scale microbial patterns?
While it is too early to study how the microbial biogeopgraphy in the MPAs may change, we could address these questions and evaluate if variation in microbial composition and diversity is associated with bottom trawling activity in the North Sea. In fact, this would be a good start and allow to develop a baseline understanding of how trawling may shape benthic microbial patterns in the North Sea.
With these questions in mind, we endeavored on two expeditions to collect sediment samples across a regional scale in the North Sea (~hundreds of kilometers, Figure 1). Offshore field collections can be challenging. The area is large and bad weather conditions may make your field trip difficult or at least unpleasant. With the Research Vessel Heincke of the Alfred Wegener Institute, its great crew and a great team of scientists, we went on two expeditions. We assembled a large dataset of sediment samples and extracted DNA to characterize with 16S rDNA microbial fingerprinting the microbial composition in these sediments. In addition, we measured sediment characteristics, bottom temperature, organic content and obtained GPS data on bottom trawling activity in the North Sea and spatial data on the intensity of natural disturbance driven by wind and currents, which may of course also affect microbial patterns.
From this dataset we could learn a great deal about the microbial biogeography of the North Sea, including patterns associated with bottom trawling activity.
Sediment characteristics are most important
Whether we looked at composition, diversity or potential metabolism, sediment grain size and mud content were always important (Figure 2, 3). This indicates that the foremost drivers of microbial communities are their local environment. The median grain size, for instance, determines how permeable the sediment is and how easily seawater may move in and out. The mud content also affects the permeability, but may also enhance the formation of micro-environments, providing unique conditions for microbes.
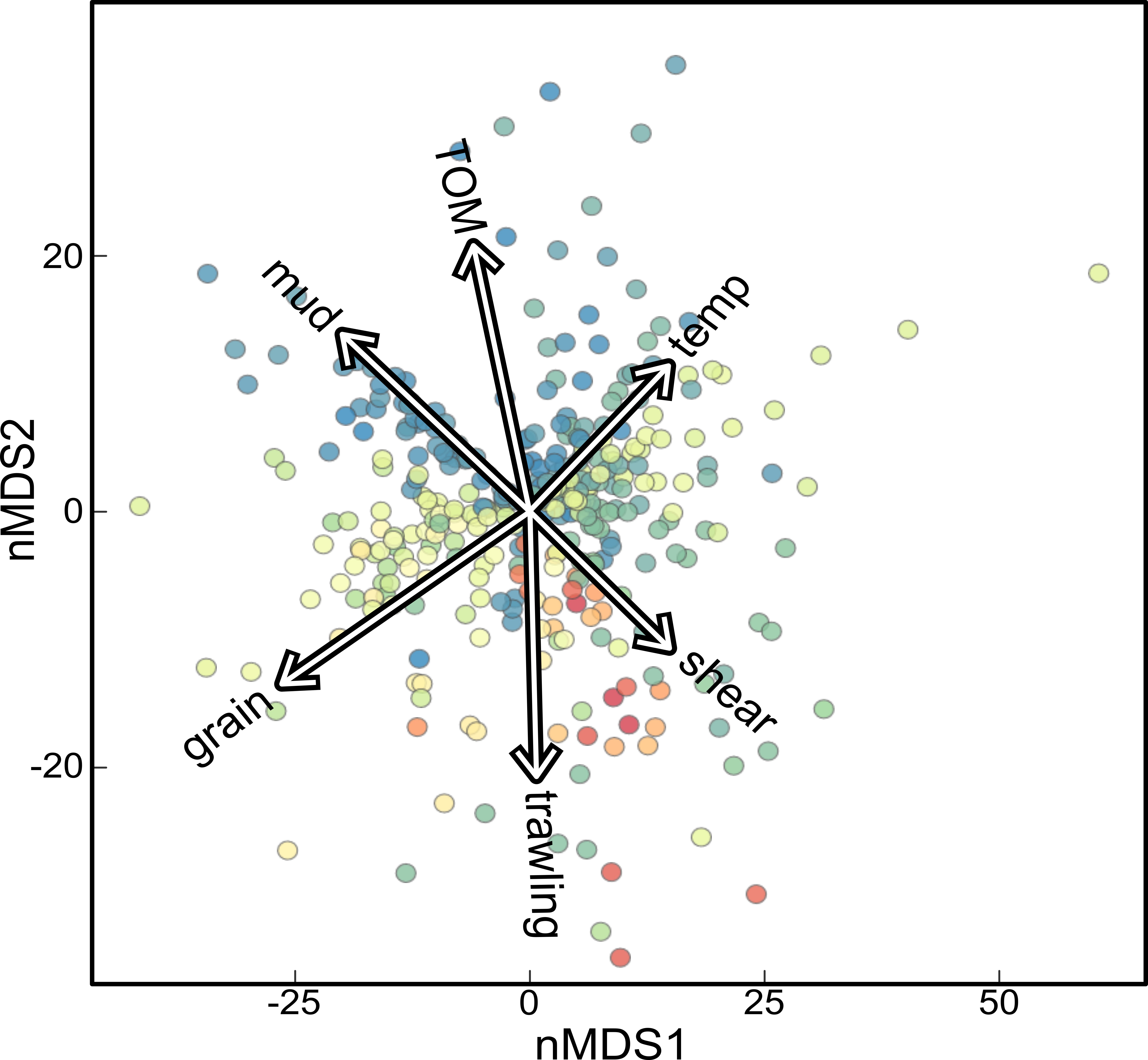
are clustered based on similarity in their microbial
communities. The vectors indicate how environ-
mental parameters are associated with the data.
Natural disturbance and anthropogenic disturbance matter both
Besides the properties of the sediment, we also found community composition and diversity to be associated with natural disturbance and trawling effort. The effects on microbial composition seemed to partially overlap (Figure 2), which could suggest that both forms of disturbance may have similar effects on sediment microbiota. However, this overlap was only partial which indicates the effects of natural bottom disturbance and trawling are not identical.
On microbial diversity, however, the effects of natural bottom disturbance and trawling were rather opposite. While diversity increased with natural disturbance, it declined or yielded more complex relationships with trawling effort (Figure 3). While it is difficult to interpret what changes in microbial diversity may entail to the ecosystem, these trends are noteworthy and suggest that disturbance levels, including those of bottom trawling, may influence microbial communities in different but structural ways.
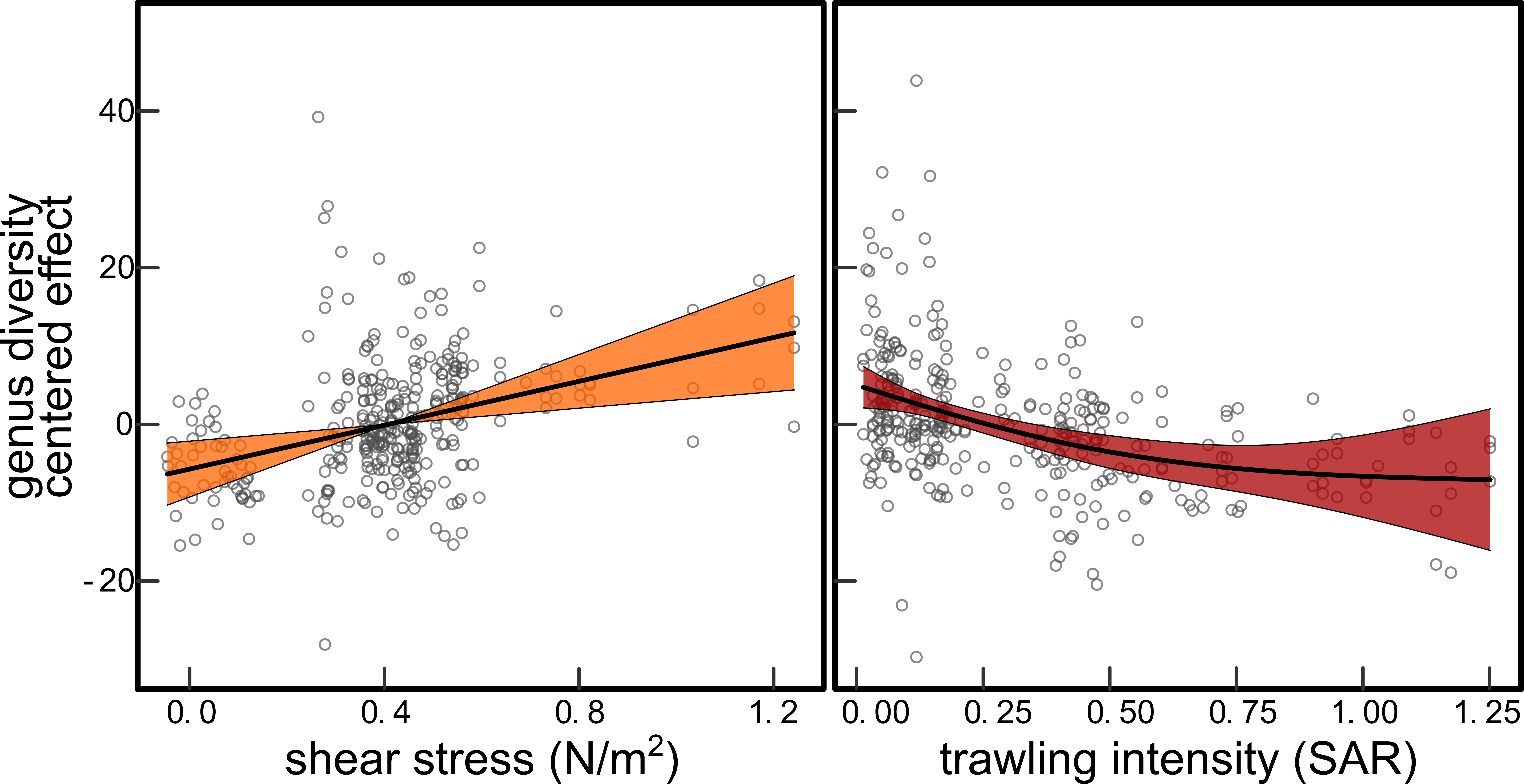
microbial diversity at the genus level in the North Sea
is associated with natural disturbance (shear stress
in N/m2) and bottom trawling intensity (in swept area
ratio per year).
Potential microbial metabolism yields also a bottom trawling signal
Finally, our data reflect that such changes in composition and diversity may come with changes in microbial metabolism, related to the consumption of oxygen or organic nitrogen. These data are only based on metabolic properties predicted from the abundances of the microbial taxa that we detected and we did not measure chemical fluxes in the field to provide strong evidence for these patterns, which remains to be done in future study. Nonetheless, these results provide important new insights into the potential consequences of bottom trawling and hint at potential changes at a functional .
In summary
Our study provides insights into how microbial communities are shaped by their environment and how they vary throughout a large section of the North Sea. While we learned that the type of sediment is most important, we also found that microbial composition and diversity are associated with disturbance intensity, both from natural sources (driven by waves and currents) and from bottom trawling activity. These results may indicate that human activities such as bottom trawling could alter the composition and diversity of microbes. While the association between bottom trawling and microbial biogeography merits further research, this is a trend existing at a large scale (hundreds of kilometers) and it is important that we have uncovered it.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in