"Dude, what's in your SynCom?"
Published in Microbiology

Most microbiologists work with bacteriophages, albeit indirectly. Outnumbering all other entities on the planet1, bacterial viruses (phages) exist not only as free-floating viral particles but also lie hidden as prophages within the genomes of most bacteria. Far from dormant, many of these prophages can undergo a process known as induction, in which they replicate and burst out of their host bacteria as infective viruses in search of new hosts. Although prophages are increasingly recognized as important players in microbial communities2, 3, very little is known about the scale and importance of induction in environmental bacteria.
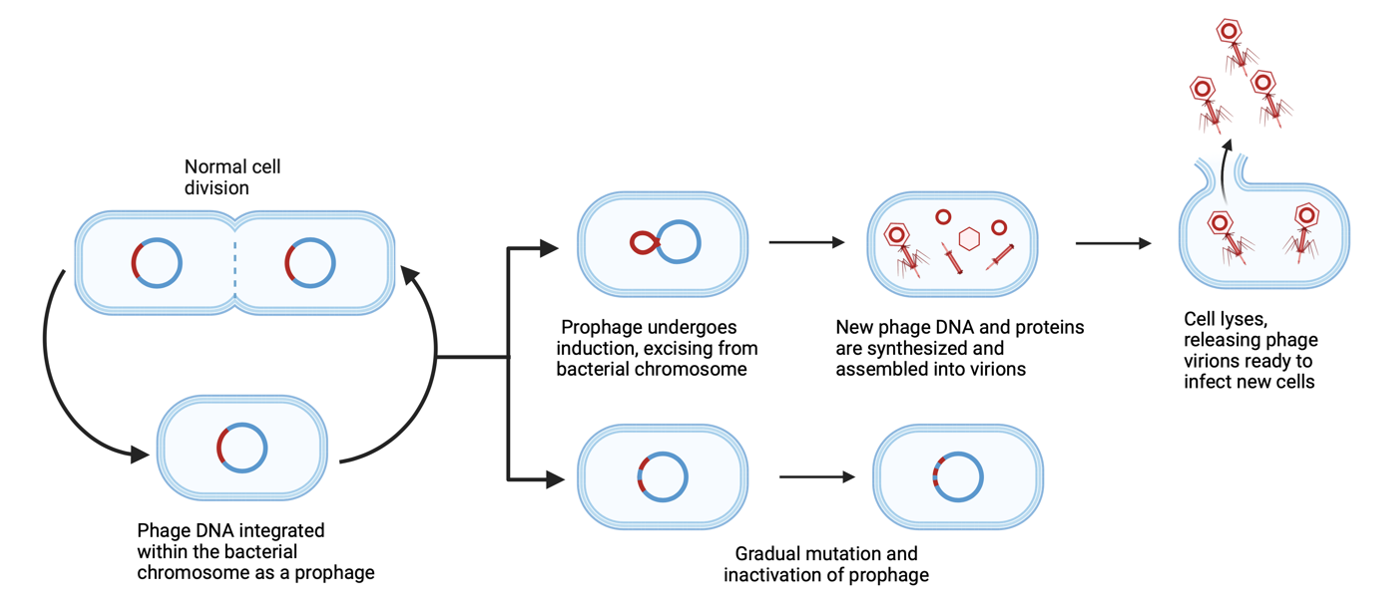
When integrated, prophages can increase host fitness by providing protection from infection by similar phages4 and may carry fitness-boosting genes such as toxins5. However, prophages are not altruistic, and those that undergo induction kill their bacterial host. This is common even in pure lab cultures, where bacteria can produce more than 109 phages/mL spontaneously6. Given the ubiquity of prophages in bacterial genomes, it is a good bet that your favorite environmental bacteria, even in pure culture, is surrounded by a cloud of induced active viruses.
Although prophage induction may not matter much when isolated in culture, it can be game-changing when different bacterial strains meet. When induced, prophages may act as bacterial weapons, infecting and killing closely related rival bacteria that were not previously infected and immunized7. In nature, encounters between different bacteria are the norm. In recognition of this, researchers are increasingly co-culturing bacteria in synthetic communities (SynComs) to create beneficial plant microbiomes8. Despite the inevitable presence of prophages in both natural and synthetic communities, the importance of their induction is rarely considered.
Here, we investigated prophage activity and diversity in bacterial isolates from the flag leaves of wheat grown in a single field. As part of the MATRIX project (Microbiome Assisted Triticum Resilience In X-dimensions), we and our collaborators aim to improve crop resilience using microbiome-assisted approaches. Interested in the importance of microdiversity within bacterial species, we decided to study prophages in bacterial isolates from the wheat phyllosphere.
In this study, we investigated the prophage content of 63 Erwinia and Pseudomonas bacterial isolates from wheat flag leaves grown in the same field. To identify and quantify all active prophages within these strains, we developed and validated Virion Induction Profiling Sequencing (VIP-Seq). Using VIP-Seq, we discovered 120 prophages spontaneously induced in liquid culture, precisely identifying their genomic positions and quantifying their concentrations.
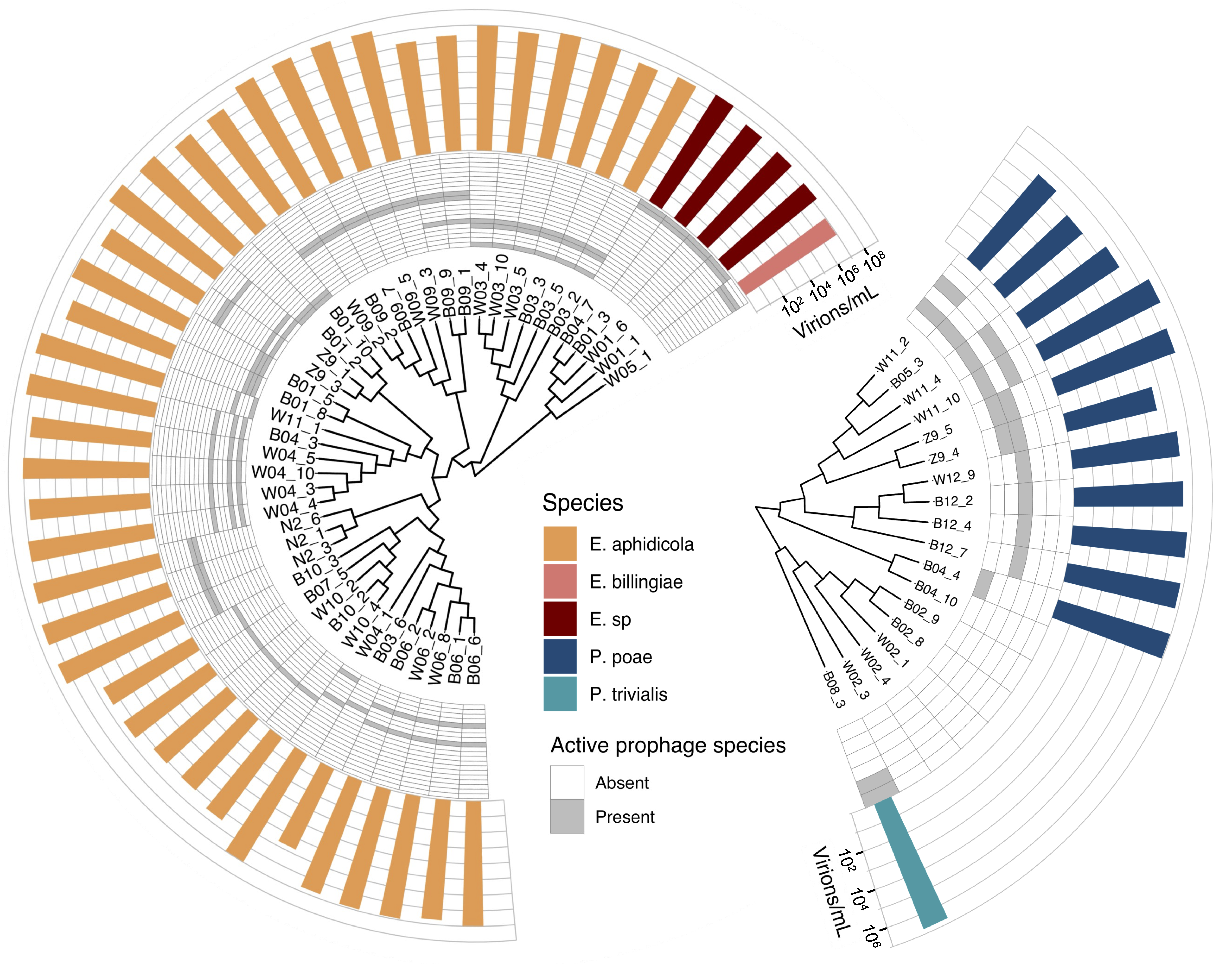
Figure 2. Genetic similarity of 45 Erwinia and 18 Pseudomonas strains. Also shown are the active prophages in each strain at species-level clusters. Finally, the aggregated virion concentrations in overnight culture for each strain is shown in a bar-plot.
Many of the Erwinia strains spontaneously produced very high concentrations of induced prophage (>108 virions/mL), while the Pseudomonas strains produced somewhat lower concentrations (~106 virions/mL). The 120 active prophages bore little similarity to known phages, and were clustered in 28 novel species clusters, which we named after Norwegian mountains. We were surprised to find such extensive prophage diversity (and activity!) within very similar bacteria isolated from a single wheat field. This was particularly true for the 40 E. aphidicola isolates. To quantify this diversity, we conducted a core genome analysis and found that most of the genetic diversity within the E. aphidicola isolates was encoded by active prophages. Clearly, prophages can be important mediators of bacterial diversity.
In addition to contributing genetic diversity, prophages are also potential bacterial weapons. To test this, we challenged all Erwinia strains with the induced prophages from all other Erwinia strains. Unexpectedly, we found many induced prophages could kill rival bacteria, with 22% of all challenges resulting in phage infection. Many of the Erwinia strains also had CRISPR spacers targeting prophages from rival strains, supporting the hypothesis that prophages are an active threat in the phyllosphere and may mediate warfare between bacterial strains.

with the spontaneously induced prophages of rival Erwinia strains.
Plaques indicate successful infection by induced prophages.
Although these results are certainly interesting, a lab culture is very different to a wheat leaf. Are prophages similarly active on leaves? As a proof of concept, we monitored prophage induction in a simple plant model by spraying wheat seedlings with an E. aphidicola strain and monitoring bacterial and phage populations over the next five days. During this time, the population of induced prophages increased significantly, demonstrating high levels of prophage induction in planta. However, the bacterial strain could not stably colonize the wheat seedlings, limiting further experiments. We hope to further investigate prophage induction in planta in future studies, perhaps using bacteria which more easily colonize seedlings.
We summarize our key takeaways as follows:
- Erwinia and Pseudomonas strains from the wheat phyllosphere contain diverse collections of prophages that greatly contribute to bacterial diversity. Many of these are active and spontaneously induced at high titres.
- The identified Erwinia prophages are highly active against rival Erwinia strains, suggesting they may mediate warfare between rival bacterial strains of the same species in the phyllosphere.
- Our VIP-Seq protocol accurately identifies active prophages in bacterial isolates and quantifies their induced titres.
Furthermore, we argue these results are of general importance to our understanding of microbial communities. Demonstrating high levels of prophage induction both in vitro and in planta, we show prophage-mediated warfare may modulate competition between bacterial strains. This is particularly relevant to the study of SynComs in plant science, where numerous bacterial strains (and their induced prophages) are combined to create beneficial plant microbiomes. Here, we show these viruses can have a dramatic effect on other members of the microbial community. If SynComs are to reliably improve crop fitness, we need to profile their resident prophages. While some of these may kill pathogenic strains and promote the establishment of beneficial communities, others may eliminate desirable plant-beneficial bacteria or spread harmful genes. Regardless, they should not be ignored. We hope our results convince others of the importance of these fascinating viruses!
Bibliography
- Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011; 1:31-45.
- Henrot C, Petit MA. Signals triggering prophage induction in the gut microbiota. Mol Microbiol. 2022; 118:494-502.
- Lugli GA, Milani C, Turroni F, Tremblay D, Ferrario C, Mancabelli L, et al. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol. 2016; 18:2196-213.
- Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, et al. Prophages mediate defense against phage infection through diverse mechanisms. Isme J. 2016; 10:2854-66.
- Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. Isme J. 2017; 11:1511-20.
- Owen SV, Wenner N, Canals R, Makumi A, Hammarlof DL, Gordon MA, et al. Characterization of the Prophage Repertoire of African Salmonella Typhimurium ST313 Reveals High Levels of Spontaneous Induction of Novel Phage BTP1. Front Microbiol. 2017; 8:235.
- Li XY, Lachnit T, Fraune S, Bosch TCG, Traulsen A, Sieber M. Temperate phages as self-replicating weapons in bacterial competition. J R Soc Interface. 2017; 14.
- Morella NM, Gomez AL, Wang G, Leung MS, Koskella B. The impact of bacteriophages on phyllosphere bacterial abundance and composition. Mol Ecol. 2018; 27:2025-38.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in