The why and how of it:
Myeloid ecotropic virus insertion site 1 (MEIS1) is essential for normal hematopoiesis and is one the most dysregulated genes in acute leukemias. Previous work, including that of our own, has shown that MEIS1 acts as an important driver for leukemogenesis1 and is overexpressed in a large subset of acute myeloid leukemia (AML) patients2,3. The transcriptional regulation of MEIS1 is complex and the mechanisms of its dysregulation are largely unknown. The MEIS1 locus spans some 175kb of the genome and recent findings suggest that this large genomic region encompasses multiple regulatory regions including several enhancers4-7. In our previous work, we assayed several human leukemic cell lines with variable levels of MEIS1 expression and identified three candidate enhancer regions, located upstream, intronic and downstream of the MEIS1 gene, based on epigenetic markers6. In our current study, we set out to further characterize these enhancers in human AML cells as well as identify the key transcription factors (TFs) associated with their function.
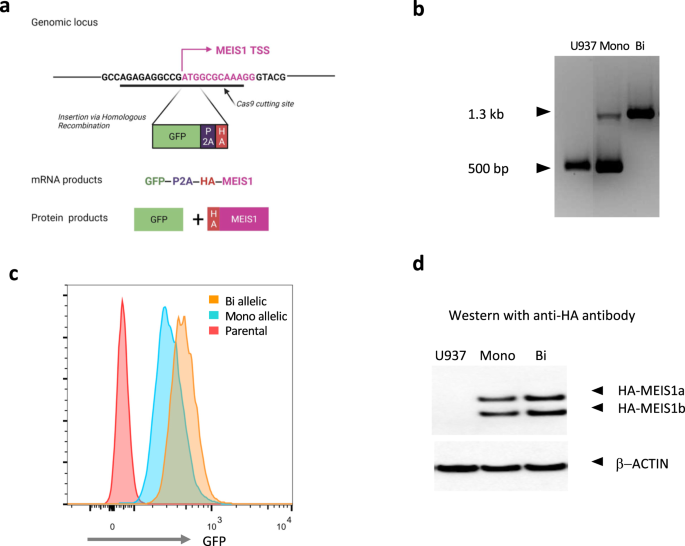
We first created a knock-in green florescent protein (GFP) reporter system at the endogenous MEIS1 locus in a human AML cell line. This reporter system allows a quick readout of MEIS1 transcription via GFP levels by flow cytometry. Our reporter construct allowed the co-transcription of GFP and HA-tagged MEIS1 using the endogenous MEIS1 transcriptional regulatory machinery. We then performed a CRISPR-Cas9 screen with 23 guide RNAs along the MEIS1 locus and looked for decrease in GFP levels as an indication of targeting of a transcriptional regulatory element. We observed the greatest GFP loss when targeting the enhancer region located in intron 6 (more than 10 Kb downstream of the transcriptional start site) which we named E2.
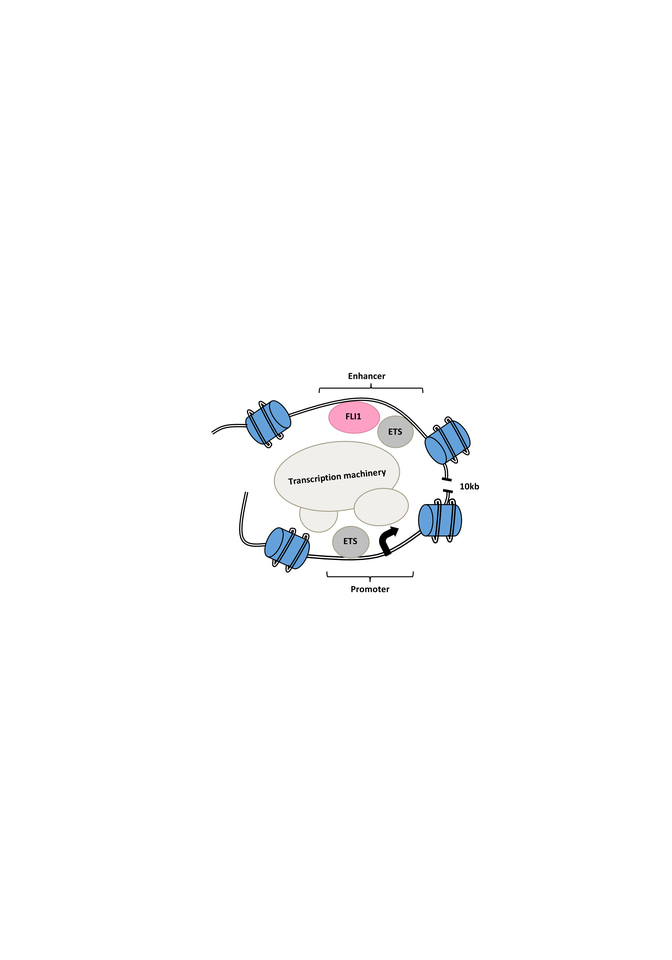
We then performed chromosome conformation capture (3C) assay and Hi-C analysis on E2 targeted GFP- cells compared to intact GFP+ cells and looked for changes in the interaction of the E2 enhancer and the promoter region as well as changes to the chromatin structure of the MEIS locus. In the E2 targeted GFP- cells, we observed less looping and interaction between the enhancer and the promoter as well as a shift to a closed chromatin structure (Figure 1).
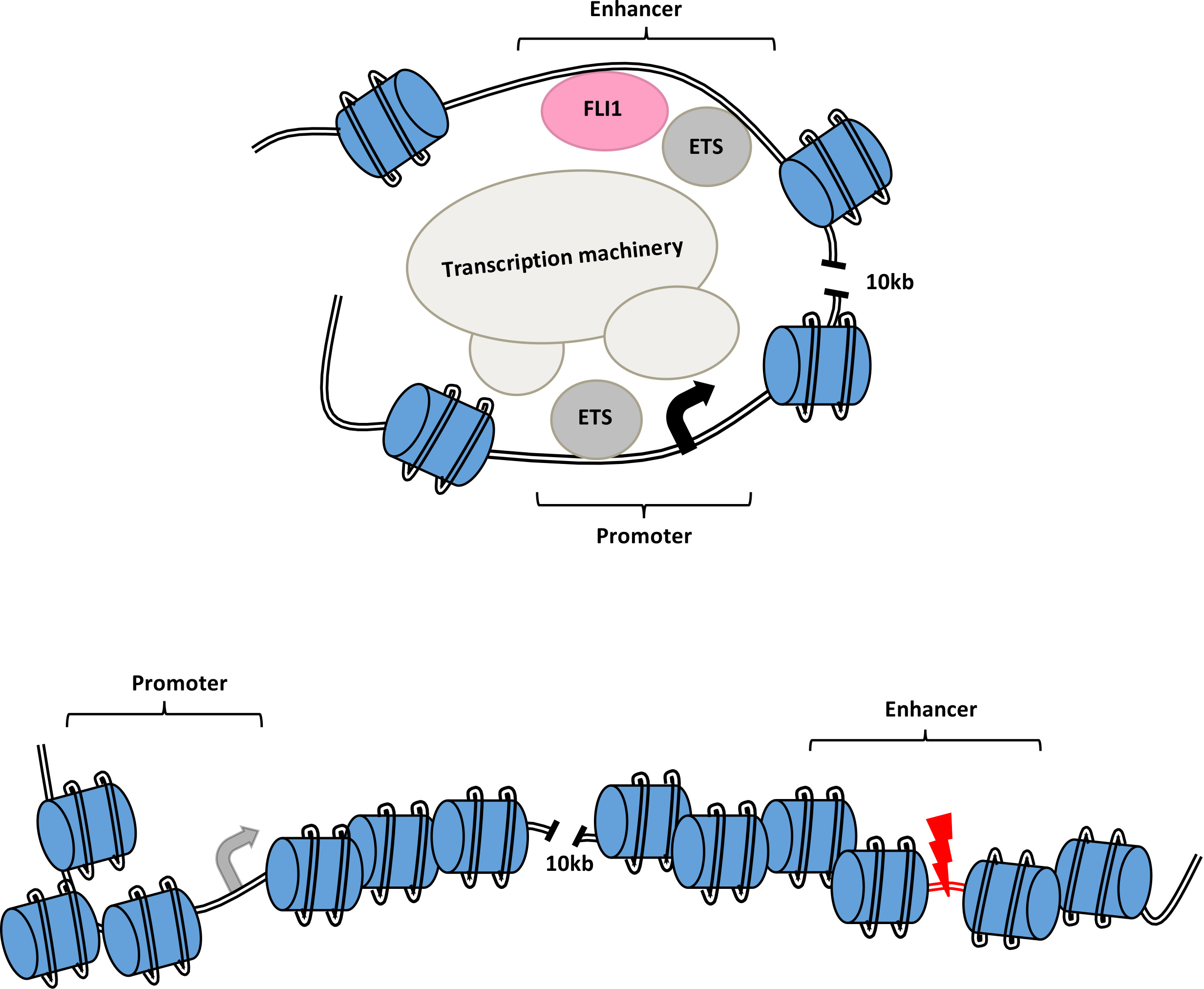
Figure 1. Schematic of open chromatin and looping interaction of the FLI1-bound intronic enhancer of the MEIS1 gene and its promoter region. Mutation of the enhancer region or targeting of FLI1 disrupts MEIS1 transcription.
To further understand the E2 function, we used in silico prediction in combination with oligo pull-down, mass-spectrometry and knockout analysis to identify important TFs for MEIS1 transcription. Our best candidate was FLI1, an E-twenty-six (ETS) transcription factor. Knocking out FLI1 via CRISPR-Cas9 resulted in the reduction in GFP and MEIS1 levels. We also observed direct binding of FLI1 to the E2 region of the MEIS1 locus in various human AML cell lines. The FLI1 binding correlated with enrichment of H3K27ac in MEIS1-high healthy and leukemic cells, suggesting an open chromatin (Figure 1). Interesting, we discovered a positive feedback loop between mouse Fli1 and Meis1, in various Hox/Meis1 induced murine leukemic models8,9, where we detected direct Meis1 binding to an intronic region of the Fli1 gene using chromatin immunoprecipitation.
We further studied the relationship between FLI1 and clinical outcome by analyzing publicly available patient datasets. High FLI1 transcript levels correlated with adverse overall survival in AML. We also observed a similar correlation for another ETS factor, ELF1, which we had previously shown to bind and upregulate MEIS1 expression in AML7, suggesting a broader unrecognized role for ETS factors in AML.
Summary and perspective:
In summary, our study revealed FLI1 as a key transcriptional regulator of MEIS1. In addition, our study expands the role of ETS factors in AML (Figure 1) and our knock-in system constitutes a feasible tool for a more detailed understanding of transcriptional regulatory elements and their interactome. We believe that our study has only scratched the surface for uncovering the complex regulatory relationship between ETS factors and MEIS1.
References
- Argiropoulos, B., Yung, E. & Humphries, R.K. Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes Dev 21, 2845-2849 (2007).
- Kuhn, M.W., et al. Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia. Cancer Discov 6, 1166-1181 (2016).
- Kumar, A.R., et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood 113, 1756-1758 (2009).
- Royo, J.L., et al. Identification and analysis of conserved cis-regulatory regions of the MEIS1 gene. PLoS One 7, e33617 (2012).
- Wang, Q.F., et al. Regulation of MEIS1 by distal enhancer elements in acute leukemia. Leukemia 28, 138-146 (2014).
- Xiang, P., et al. Delineating MEIS1 cis-regulatory elements active in hematopoietic cells. Leukemia 28, 433-436 (2014).
- Xiang, P., et al. Identification of E74-like factor 1 (ELF1) as a transcriptional regulator of the Hox cofactor MEIS1. Exp Hematol 38, 798-798, 808 e791-792 (2010).
- Schneider, E., et al. Micro-ribonucleic acid-155 is a direct target of Meis1, but not a driver in acute myeloid leukemia. Haematologica 103, 246-255 (2018).
- Schneider, E., et al. MicroRNA-708 is a novel regulator of the Hoxa9 program in myeloid cells. Leukemia 34, 1253-1265 (2020).
Follow the Topic
-
Leukemia

This journal publishes high quality, peer reviewed research that covers all aspects of the research and treatment of leukemia and allied diseases. Topics of interest include oncogenes, growth factors, stem cells, leukemia genomics, cell cycle, signal transduction and molecular targets for therapy.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in