Evaluation of DNA methylation biomarkers ASCL1 and LHX8 on HPV-positive self-collected samples from primary HPV-based screening
Published in Cancer
Primary cervical screening with high-risk human papillomavirus (HPV) testing is more effective in protecting against cervical cancer and precancer (CIN3+) than cervical cytology tests (1, 2). An advantage of HPV testing is that it can be performed on clinician-collected as well as self-collected screening samples. Self-sampling is a promising strategy to overcome barriers to cervical screening and to increase screening coverage (3). However, current triage methods like cytology, nowadays require recalling of women who are HPV-positive on a self-collected sample for clinician-based sampling. Unfortunately, not all women adhere to this advice and a part does not show up at the clinician’s office. Therefore, alternative triage methods that can be directly applied to self-collected samples, are highly warranted.
DNA methylation of host-cell genes has great potential as alternative triage strategy for HPV-based cervical screening. This approach can specifically detect advanced cervical lesions that require treatment (4), with the potential of being directly applicable to self-collected screening samples. Several studies have reported on the effectiveness of DNA methylation analysis for direct triage in HPV self-sampling. However, most of these studies have been performed in cohorts of under screened or never-screened women and referral populations. In this study, for the first time, the triage performance of host-cell methylation analysis was evaluated in HPV-positive self-collected samples from women who were offered primary HPV self-sampling for cervical cancer screening.
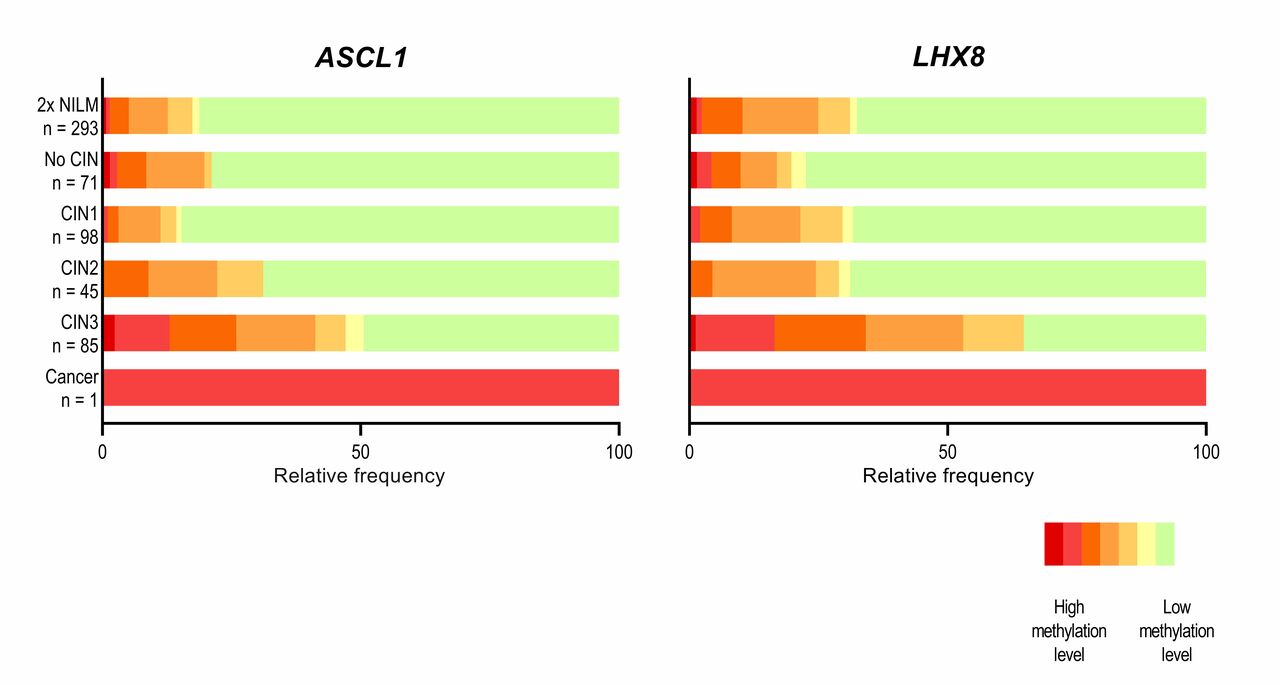
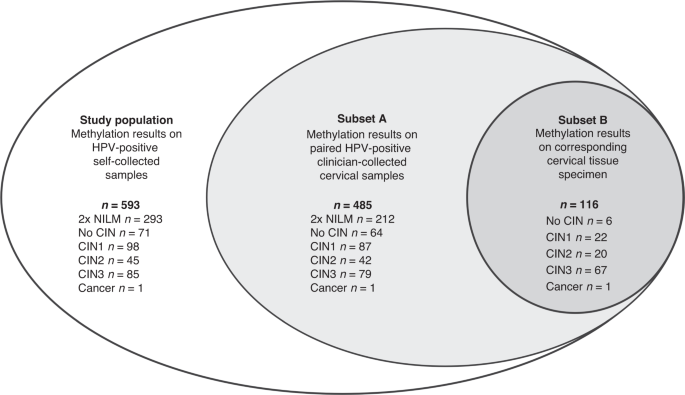
Our study focuses on evaluating two methylation markers, ASCL1 and LHX8, that were discovered in self-collected samples (5). These markers were previously analysed on clinician-collected cervical samples (6, 7) and self-collected samples from screening non-attendees (5). We conducted a post-hoc analysis of the IMPROVE study (Netherlands Trial Register, number NTR5078), that evaluated HPV self-sampling within the setting of the Dutch cervical screening program (8). The unique study design of the IMPROVE study allowed us to directly compare methylation analysis between self-collected and clinician-collected samples from the same HPV-positive women. We included 593 self-collected samples from HPV-positive women who participated in the IMPROVE study, consisting of 462 control women (≤CIN1), 45 women with CIN2, 85 women with CIN3 and 1 women with cervical squamous cell carcinoma. Self-samples were tested for methylation of ASCL1 and LHX8 using quantitative multiplex methylation-specific PCR. For comparison, we also had available the ASCL1/LHX8 methylation data on paired HPV-positive clinician-collected cervical samples for 485 women (7).
In our study, we found that the methylation levels of ASCL1 and LHX8 in self-collected samples of HPV-positive women increased as the severity of underlying cervical disease worsened and were significantly higher in women with CIN3 + than in control women with no evidence of disease. The ASCL1/LHX8 marker panel showed a sensitivity of 73.3% (95%CI 63.9-82.6%) for detecting CIN3+, with a corresponding specificity of 61.1% (95%CI 56.9-65.4%). The triage performance of ASCL1/LHX8 methylation analysis on self-collected samples was somewhat lower than that on clinician-collected cervical samples, with a relative sensitivity for CIN3 + detection of 0.95 (95% CI 0.82–1.10) and a relative specificity of 0.82 (95% CI 0.75–0.90). Our data indicate that the methylation marker panel ASCL1/LHX8 constitutes a feasible direct triage method for the detection of CIN3 + in HPV-positive women participating in routine screening by self-sampling. The advantage of DNA methylation analysis as a triage test is the use of an objective, non-morphological assay, with a high reproducibility, directly applicable to self-collected samples. Our results support further clinical validation in prospective screening studies using HPV self-sampling.
References
- Arbyn M, Ronco G, Anttila A, Meijer CJ, Poljak M, Ogilvie G, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine. 2012;30 Suppl 5:F88-99.
- Ronco G, Dillner J, Elfström KM, Tunesi S, Snijders PJ, Arbyn M, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet (London, England). 2014;383(9916):524-32.
- Serrano B, Ibáñez R, Robles C, Peremiquel-Trillas P, de Sanjosé S, Bruni L. Worldwide use of HPV self-sampling for cervical cancer screening. Preventive medicine. 2022;154:106900.
- Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14(6):395-405.
- Verlaat W, Snoek BC, Heideman DAM, Wilting SM, Snijders PJF, Novianti PW, et al. Identification and Validation of a 3-Gene Methylation Classifier for HPV-Based Cervical Screening on Self-Samples. Clin Cancer Res. 2018;24(14):3456-64.
- Dick S, Verhoef L, De Strooper LM, Ciocănea-Teodorescu I, Wisman GBA, Meijer CJ, et al. Evaluation of six methylation markers derived from genome-wide screens for detection of cervical precancer and cancer. Epigenomics. 2020;12(18):1569-78.
- Verhoef L, Bleeker MCG, Polman N, Steenbergen RDM, Meijer C, Melchers WJG, et al. Performance of DNA methylation analysis of ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST for the triage of HPV-positive women: Results from a Dutch primary HPV-based screening cohort. International journal of cancer. 2022;150(3):440-9.
- Polman NJ, Ebisch RMF, Heideman DAM, Melchers WJG, Bekkers RLM, Molijn AC, et al. Performance of human papillomavirus testing on self-collected versus clinician-collected samples for the detection of cervical intraepithelial neoplasia of grade 2 or worse: a randomised, paired screen-positive, non-inferiority trial. Lancet Oncol. 2019;20(2):229-38.
Follow the Topic
-
British Journal of Cancer

This journal is devoted to publishing cutting edge discovery, translational and clinical cancer research across the broad spectrum of oncology.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in