Expanding and consolidating the RNA-targeting CRISPR-Cas toolbox
Published in Microbiology, Protocols & Methods, and Cell & Molecular Biology

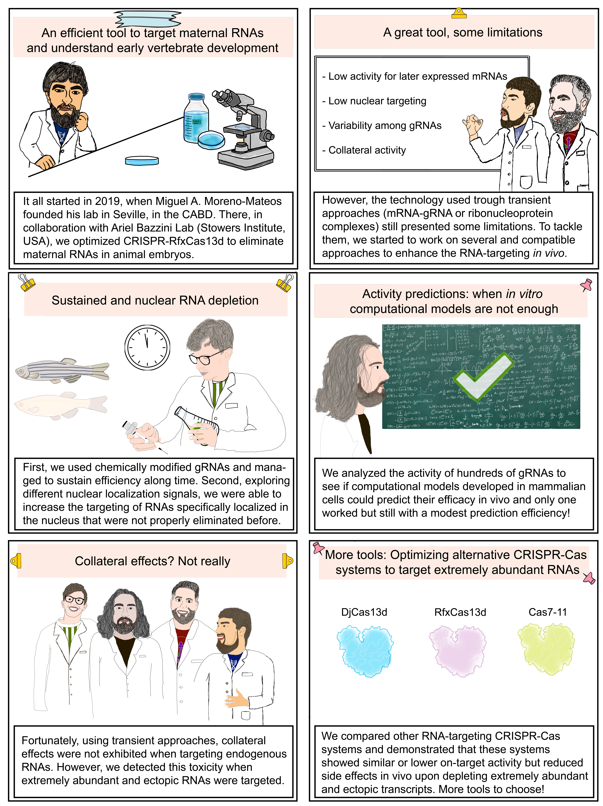
Everything started in 2019 when Miguel A. Moreno-Mateos founded its laboratory in the Andalusian Centre of Developmental Biology in Seville (Spain), with the idea of implementing new molecular technologies to systematically eliminate maternally provided mRNAs in zebrafish. In collaboration with Gopal Kushawah and Ariel A. Bazzini from Stowers Institute for Medical Research in Kansas (MO, USA), we optimized CRISPR-RfxCas13d (Konnerman S. et al., 2018) in zebrafish and other animal embryos demonstrating that CRISPR-RfxCas13d was cost-effective, specific and efficient to eliminate mRNAs in vivo using mRNA-gRNA or ribonucleoprotein (RNP) formulations (Kushawah G. et al., 2020; Hernández-Huertas L. et al., 2022).
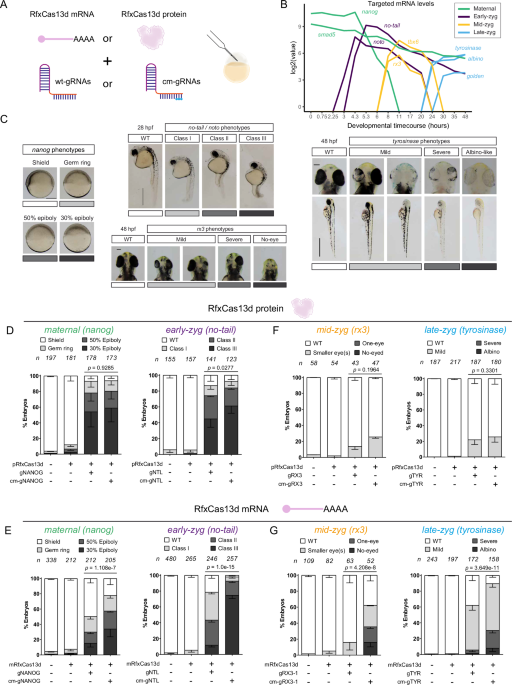
During the following years, we have performed several screenings using our optimized CRISPR-RfxCas13d system. For example, we have targeted a subset of mRNAs coding kinases and phosphatases with an unknown role in early zebrafish development uncovering new factors that control the transcriptional activation of the genome after oocyte fertilization (Hernández-Huertas L. et al., 2024). However, we realized that CRISPR-RfxCas13d system presented some limitations that we wanted to address. First, we did not achieve successful results when targeting mRNAs expressed later during development. For this, in collaboration with Joan Galcerán and Ángela Nieto from Neuroscience Institute (CSIC, Alicante, Spain) and John A. Walker II and Kevin Holden from Synthego Corp. (Redwood City, CA, USA), we made CRISPR-RfxCas13d more effective and managed to sustain the RNA targeting activity for a longer time by using chemically modified gRNAs together with RfxCas13d mRNA (Fig. 1A). Similarly, by screening different nuclear localization motives we were able to identify one that enhances CRISPR-RfxCas13d RNA targeting in the nucleus (Fig. 1B) that is required for those RNAs that are mainly or only located there such as primary miRNAs or lncRNAs.
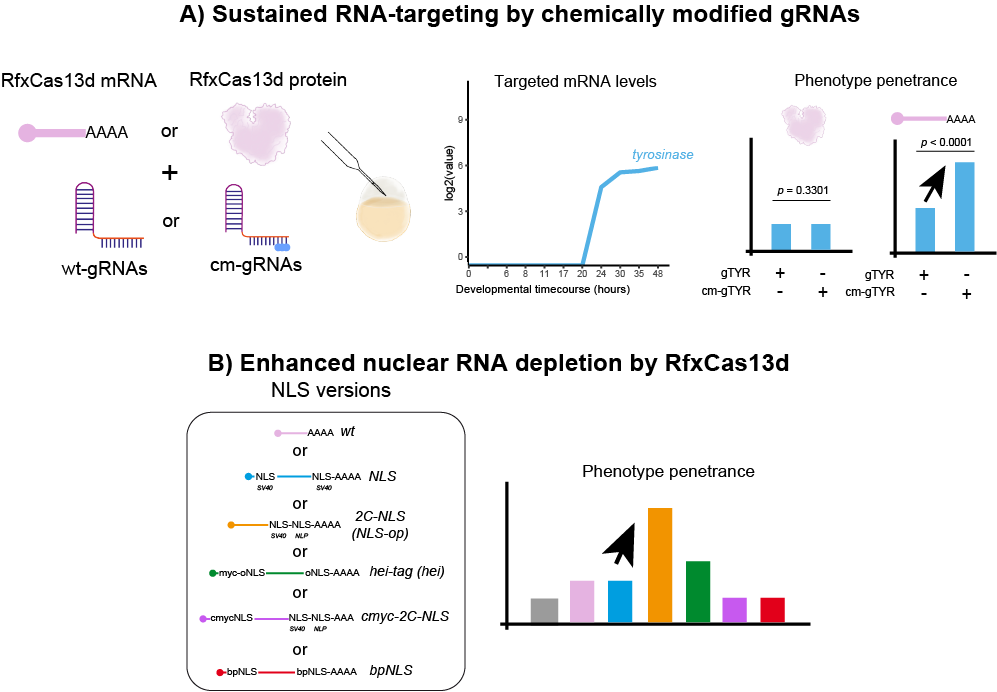
Figure 1. Sustained RNA-targeting by chemically modified gRNAs and enhanced nuclear RNA depletion in zebrafish.
Another issue that we and other colleagues had found working with RfxCas13d in animals was the variability of the system’s efficiency depending on the gRNA used. Therefore, as a pilot experiment, we analyze the activity of hundreds of gRNAs to interrogate existing computational models developed in mammalian cells for their performance predicting CRISPR-RfxCas13d activity in vivo (Fig. 2; Cheng X. et al., 2023; Wei J. et al., 2023; Wessels H. et al., 2024) in collaboration with Pedro M. Martínez-García (CABD, CSIC-UPO) and Rhonda Egidy and Anoja Perera (Stowers Institute for Medical Research, Kansas City, MO, USA). These computational models have been generated from data where Cas and gRNAs were expressed constitutively. However, we use transient approaches, employing a limited amount of gRNA and Cas that is decreasing along time. We uncovered that one of these computational models, RNAtargeting (Wei J. et al., 2023), can be used to predict gRNA efficiency when employing transient applications, though it isn’t as precise as in mammalian cells experiments.
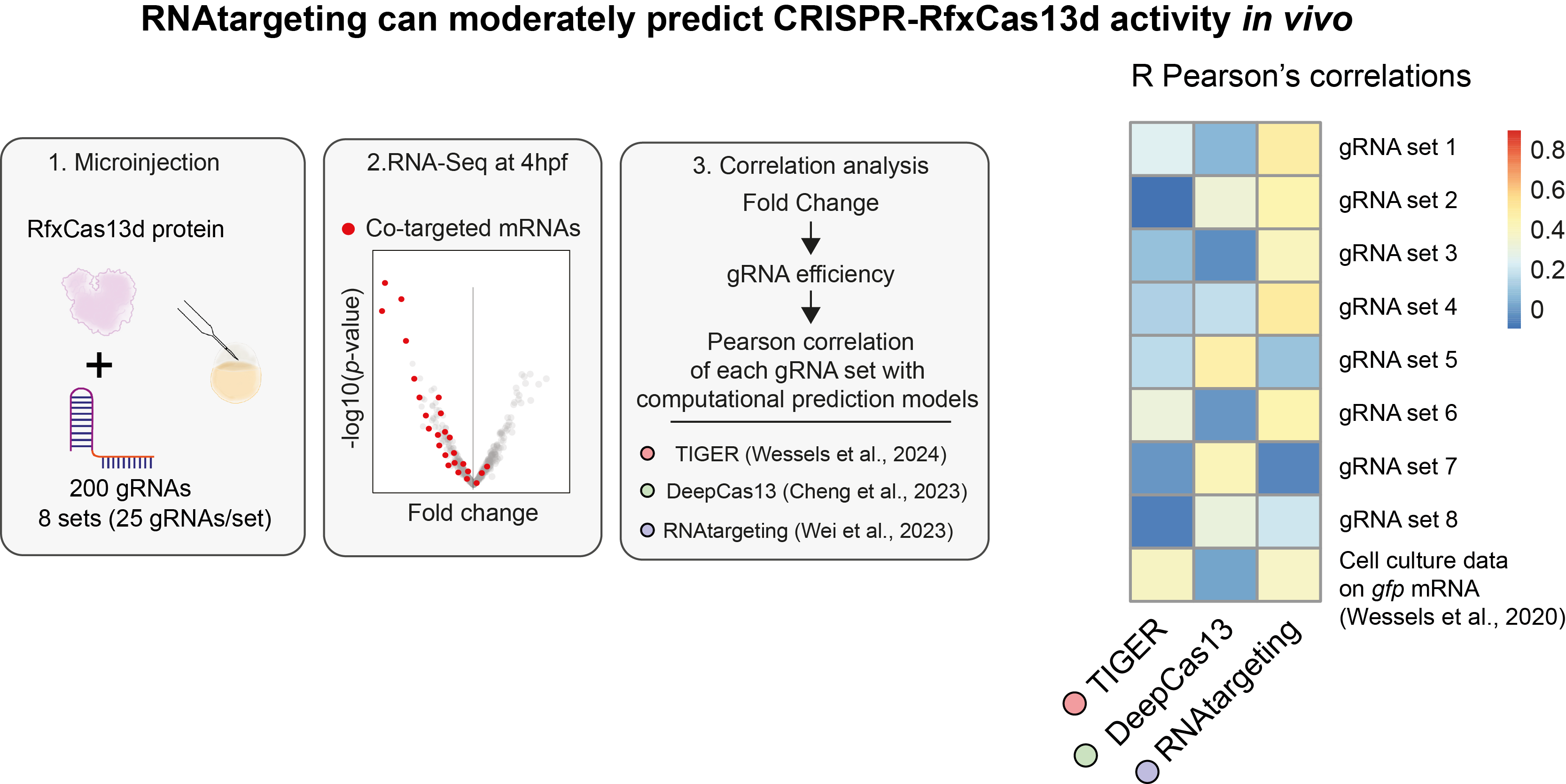
Figure 2. RNAtargeting can moderately predict CRISPR-RfxCas13d activity.
Additionally, we had also noticed that some gRNAs in vitro synthetized (IVTed) can trigger toxic effects in zebrafish embryos. Importantly, this toxicity triggered by IVTed gRNAs is completely resolved when using chemically synthetized gRNAs. Since they are expensive, Anthony J. Treichel, Gopal Kushawah and Gabriel da Silva Pescador from Ariel A. Bazzini lab, generated an in vitro assay to screen for cost-effective IVTed gRNAs to discard those gRNAs that could induce toxic effects (Fig. 3). Why some particulars IVTed gRNAs induce this toxicity is something that remains to be further analyzed.
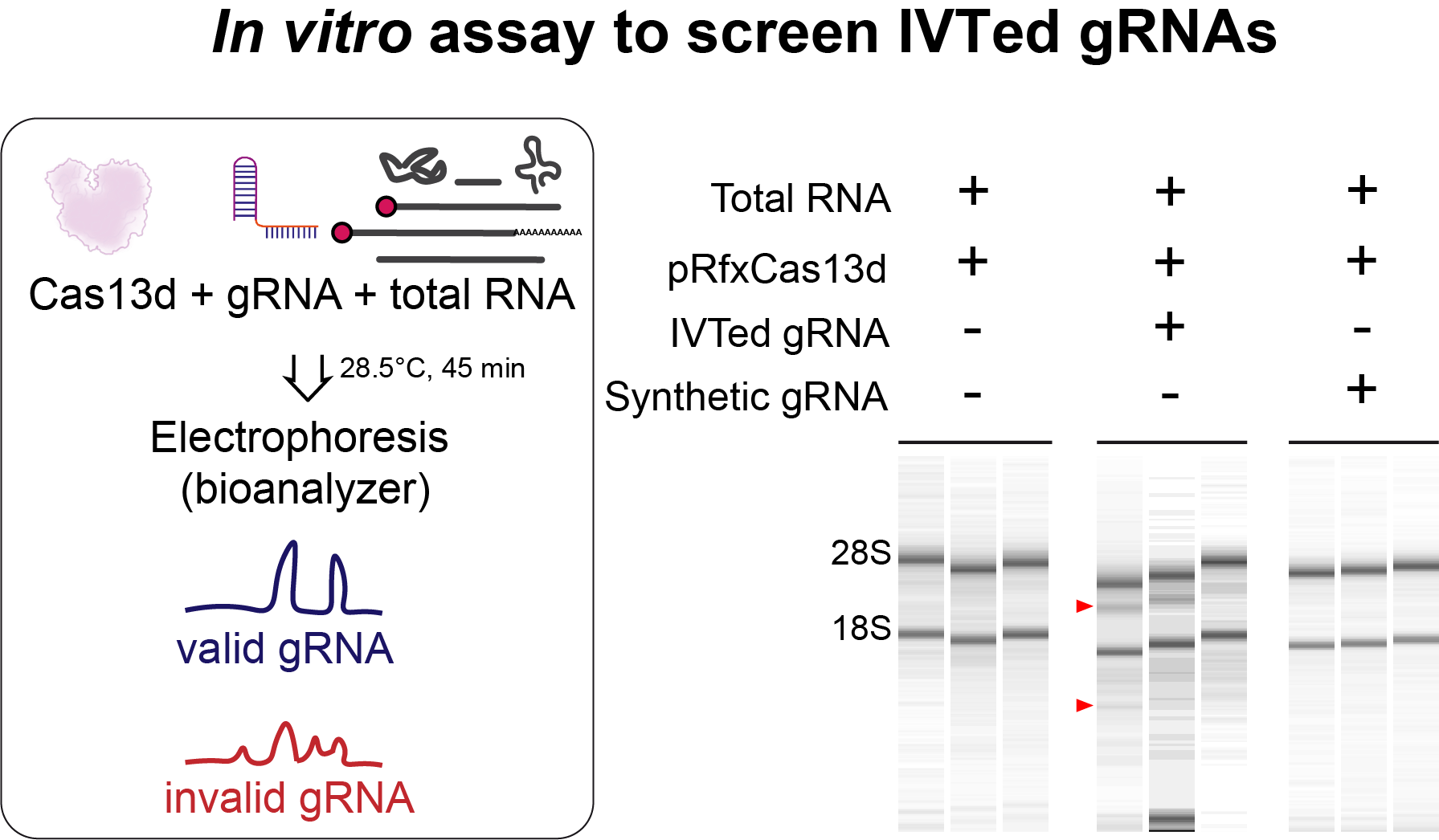
Figure 3. In vitro assay to screen for IVTed gRNAs.
But essentially, the most striking section of this paper and what made us to revise all our previous data from CRISPR-RfxCas13d experiments was the so-called collateral activity. Until 2022, it had not been reported collateral activity from CRISPR-RfxCas13d activity in cell culture experiments or in vivo applications. When these reports were published (Ai Y. et al., 2022; Kelley C. et al., 2022; Li Y. et al., 2023; Shi P. et al., 2023), the whole lab led by Ismael Moreno-Sanchez, Luis Hernandez Huertas and Daniel Nahón-Cano, and with the invaluable help of Alejandra Cano-Ruiz, as a cooperative and team work, rapidly revisited all our previous experiment with zebrafish embryos. We were unable to observe collateral activity using our optimized and transient approaches. However, we were convinced that under certain circumstances these collateral effects could be detected in zebrafish embryos. The hallmarks of this side effect in mammalian cells are i) a reduction of cell proliferation, ii) a decay of other RNAs apart from targeted RNA and iii) a degradation of 28S ribosomal subunit RNA (rRNA). Using a double reporter system, we could see that when CRISPR-RfxCas13d targeted highly expressed ectopic reporter mRNA led to collateral activity.
Importantly, CRISPR-RfxCas13d targeting endogenous and very abundant mRNAs did not significantly cause any of these hallmarks rather than a weak but still detectable 28S rRNA degradation (Fig. 4A). Therefore, we could demonstrate that our optimized CRISPR-RfxCas13d approaches were generally safe, but scientists need to be cautious when using it on extremely expressed RNAs.
For that particular situation, together with Carlos Gómez-Marín from Manuel J. Muñoz lab (CADB, UPO) and Laura Tomás-Gallardo and Alejandro Díaz-Moscoso from Proteomics and Biochemistry Platform (CABD, UPO-CSIC), we have implemented two other RNA-targeting CRISPR-Cas systems to be used as RNP in zebrafish embryos, CRISPR-Cas7-11 (Özcan A. et al., 2021) and CRISPR-DjCas13d (Wei J. et al., 2023). We compared them together with a high-fidelity version of RfxCas13d for on-target and collateral activity (Fig. 4B). We concluded that DjCas13d had fewer collateral activity than RfxCas13d while maintaining similar efficiency and Cas7-11 had no detectable side effects although lower on-target activity. This on-target activity was much lower using high-fidelity version of RfxCas13d recapitulating a recent report in mammalian cells (Hart et al., 2025).
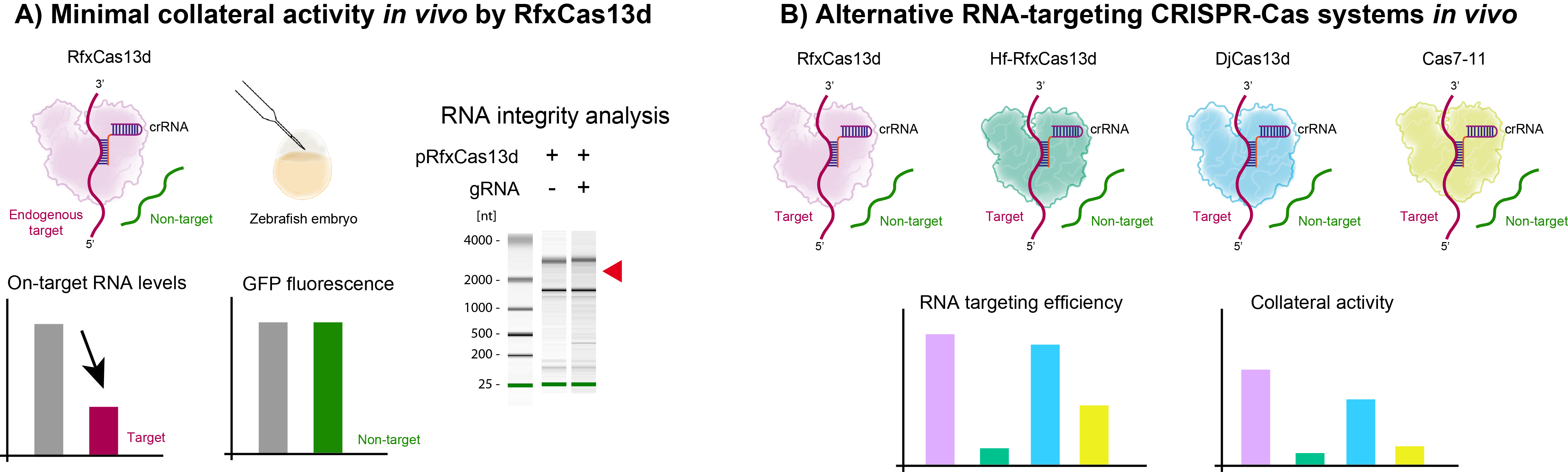
Figure 4. Minimal collateral activity in vivo by RfxCas13d and alternative RNA-targeting CRISPR-Cas systems.
We believe that our study will contribute to improve RNA-editing technology in vivo by making CRISPR-Cas13 more precise, efficient and accessible. These advances could eventually help in biomedical research, biotechnology, and even future treatments for diseases that involve defective RNA.
References
Konermann, S. et al. Transcriptome engineeringwith RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018).
Kushawah, G. et al. CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev. Cell 54, 805–817.e7 (2020).
Hernández-Huertas, L. et al.Optimized CRISPR-RfxCas13d system for RNA targeting in zebrafish embryos. STAR Protoc. 3, 101058 (2022).
Hernández-Huertas, L. et al. CRISPR-RfxCas13d screening uncovers Bckdk as a post-translational regulator of the maternal-to-zygotic transition in teleosts. bioRxiv, https://doi.org/10.1101/2024.05.22.595167 (2024).
Cheng, X. et al. Modeling CRISPR-Cas13d on-target and off-target effects using machine learning approaches. Nat. Commun. 14, 1–14 (2023).
Wei, J. et al. Deep learning and CRISPR-Cas13d ortholog discovery for optimized RNA targeting. Cell Syst. 14, 1087–1102.e13 (2023).
Wessels, H. H. et al. Prediction of on-target and off-target activity of CRISPR–Cas13d guide RNAs using deep learning. Nat. Biotechnol. 42, 628–637 (2024).
Ai, Y., Liang, D. &Wilusz, J. E. CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells. Nucleic Acids Res. 50, E65 (2022).
Kelley, C. P., Haerle, M. C. & Wang, E. T. Negative autoregulation mitigates collateral RNase activity of repeat-targeting CRISPRCas13d in mammalian cells. Cell Rep. 40, 111226 (2022).
Li, Y. et al. The collateral activity of RfxCas13d can induce lethality in a RfxCas13d knock-in mouse model. Genome Biol. 24, 20 (2023).
Shi, P. et al. Collateral activity of the CRISPR/RfxCas13d system in human cells. Commun. Biol. 6, 334 (2023).
Özcan, A. et al. Programmable RNA targeting with the single protein CRISPR effector Cas7-11. Nature 597, 720–725 (2021).
Hart, S. K. et al. Precise RNA targeting with CRISPR–Cas13d. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02558-3 (2025).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in