Exploring bacterial interactions - or how to become a bacteria whisperer
Published in Microbiology

Explore the Research

In vitro interaction network of a synthetic gut bacterial community - The ISME Journal
The ISME Journal - In vitro interaction network of a synthetic gut bacterial community
In the recent decade, the microbial communities we share this planet with - and especially those we share a body with - finally reached the scientific fame they deserve. Particularly, the importance of the many different microbes that live in our guts became undeniable. Understanding the functions and underlying mechanisms microbiomes take on in their host's health and disease likely will bring modern medicine vast steps forward as we become able to treat us humans as the holobionts we are. But how can we study systems like the gut microbiome, that comprises hundreds of species, millions of cells?
In around 2010, sequencing initiatives like HMP and Mega-HIT yielded the first insights into how human microbial communities are composed – but scientists were still lacking accessible model systems for functional microbiome research in the lab. While large-scale comparative sequencing approaches make complex bacterial communities theoretically more accessible to us, mechanistic understanding still relies on the isolation and cultivation of single bacteria. To move from correlation to causation, we have to disassemble these huge and complex bacterial networks. Bacterial community model systems that are smaller in numbers and easier to handle were needed to provide researchers with the possibility to dissect the individual bacterial roles in host associated observations.
Therefore, a team around Sandrine Brougiroux and Bärbel Stecher set out to move a step further and designed a bacterial community that represents the major bacterial phyla of the mouse gut, a guided set of species that can directly reflect specific microbiome functions that provide gnotobiotic mouse models with healthier hosts. The Oligo Mouse Microbiota (OMM12) finally comprised twelve bacterial strains isolated from the murine gastrointestinal tract that were found to stably colonize gnotobiotic mice over several generations, that were fully genome sequenced and made openly available in pure culture (Brugiroux et al. 2016). Up to now, this bacterial community has been used in several studies elucidating the role of the gut microbiota in different host disease contexts, ranging from colorectal cancer to enteropathogen infection to inflammatory bowel disease. While in gnotobiotic mouse models the OMM12 proofed to be a reliable tool, working the full potential of synthetic bacterial communities requires knowing more about the individual strains’ behavior in different environmental settings and the community’s ecology. Therefore, we decided it was time to explore the metabolic potential, strain interactions and community dynamics of this specific group of bacteria.
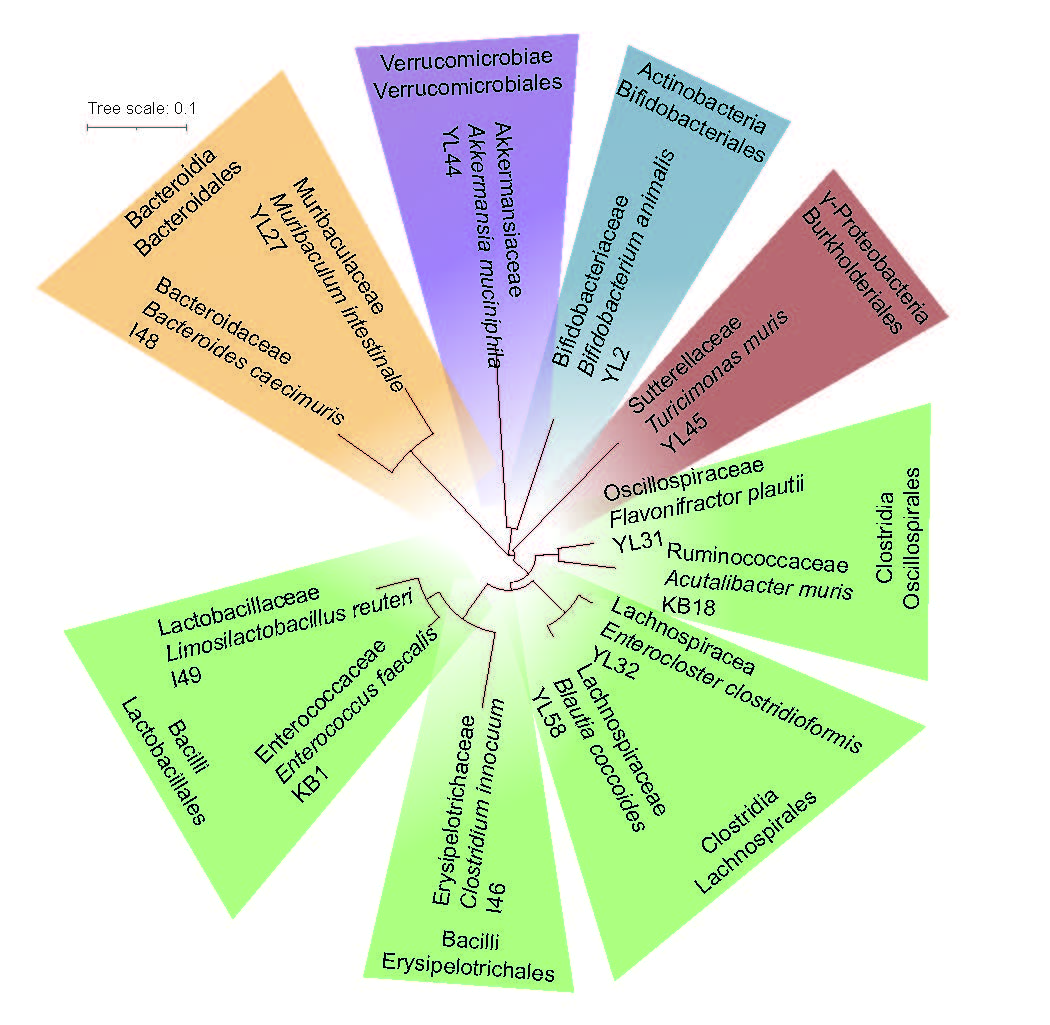
bacterial phyla of the murine gastrointestinal tract.
We started out easy and first analyzed growth behavior of all strains in monoculture in a chemically complex and nutrient rich culture medium. Further, we took samples of all twelve densely grown monocultures and took a closer look at their left-over culture medium by comparing it to fresh medium using mass spectrometry and analyzing pH values before and after growth. Thereby, we were able to determine the specific metabolic preferences of the individual bacteria and compare their metabolic profiles. To learn even more about the metabolic capabilities of the twelve OMM bacteria, we determined genome-informed metabolic models. Combining what we learned with our observations from the monoculture experiments enabled us to confirm that specific metabolic functions known to be important for gut health are realised by individual bacteria. For example, we could show that two bacteria of the consortium, Clositridum innocuum I46 and Flavonifractor plautii YL31 produce butyrate, a major energy resource for epithelial cells, or that Turicimonas muris YL45 degrades amino acids as aspartate to produce ammonia, which strongly influences the pH environment.
Besides specific metabolic consumption or production patterns, we also found quite some overlap regarding the metabolic requirements in the chosen in vitro culture medium, especially for more closely related bacterial strains. Therefore, we were interested in how this would affect bacterial interactions, as when mixed the bacteria would have to compete for the most popular nutrients. Covid-19 lockdown seemed to be a good time to screen all bacterial pairwise co-cultures (by the way it's 66), which gave first insights into assembly and ecology of the OMM12 consortium and revealed several different interaction outcomes. We found both extreme ends of the interaction spectrum, where some strain pairs just arranged with the presence of each other, resulting in neutral interaction outcomes, while others were not ready to settle the fight by competing until the very end, resulting in negative effects for both strains. But we also observed plenty of amensalistic interaction outcomes, meaning that one of the strains in a specific co-culture was affected negatively by the presence of the other strain, while the latter was neither positively nor negatively affected in its growth by the presences of the other.
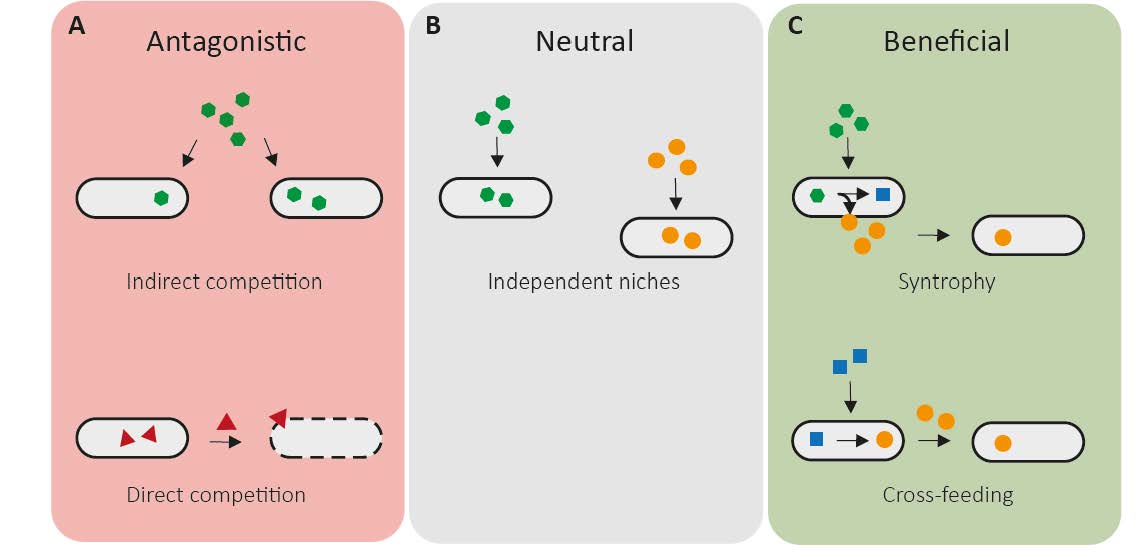
or beneficial interactions.
One interaction that stood particularly out to us was between Enterococcus faecalis KB1 and Clostridium innocuum I46. The pairwise co-culture resulted in what is classified as a “predatory” outcome, where E. faecalis strongly profited from the presence of C. innocuum, while this strain was doing pretty badly. Taking a closer look, we found that E. faecalis produces a compound, likely a bacterial toxin, that shows detrimental activity against C. innocuum, while this strain produces metabolites that its opponent can feed on. E. faecalis generally seemed to be somewhat the party pooper (pun intended) - its presences negatively affected all of the other strains of the consortium in some way or the other.
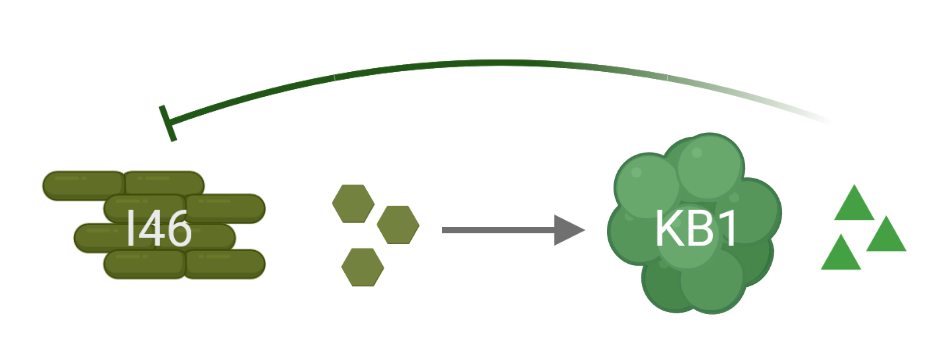
Seeing a lot of potential conflict – and from watching one or the other reality TV show, we all know this sells best – we decided to go one step further and put all the twelve strains together into the same pot. Surprisingly, this did not end up in an E. faecalis monoculture, but our bacteria somehow arranged to coexist, at least the vast majority. Still, when we dropped E. faecalis out of the consortium, not only C. innocuum seemed to be somewhat relieved. Also another strain, Bifidobacterium animalis, finally showed growth in the community when the bully was away.
However, a good bacterial parent doesn’t choose sides – informed by the bacterial metabolic capabilities we designed different culture media with specific carbon sources to feature individual strains. By that, we were able to construct bacterial communities that were dominated by Flavonifractor plautii YL31 when starch was available or Bacteroides caecimuris I48 when polysaccharides were abundant. The latter community even closely resembled what the OMM12 consortium looks like in the murine gut, leaving room for follow-up experiments comparing the communities’ behavior in vitro vs. in vivo.
Concluding, we used a bunch of different approaches ranging from mass spectrometry and genome-informed analyses for a better understanding of the individual bacterial species’ metabolisms, to measuring pairwise interactions in co-cultures, to observing bacterial ecology in community batch cultures – all to get to know a weird little group of bacteria isolated from mouse poop (Mum, Dad, are you proud?). Doing so, we provided a vast data resource, for all the people that are intrigued by the OMM12 model as well, and to be fair, that probably also rather work with twelve bacterial species than with twelve hundred. Still, we only scratched the surface of what there is to be learned about gut bacterial communities, their metabolism and their ecology - and to be honest, I can’t wait for what is there to come.
Read the full story here.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in