
Genomic analysis of bacterial communities can often be tricky, particularly in environments with many uncharacterized species. Let’s say you are interested in a specific species, because you repeatedly find its 16S rRNA sequence in your amplicon datasets and you want to learn more about its capabilities and its role in the environment. But this species does somehow not like the culturing medium you are using or the conditions you try to grow it in. So you decide to take a culture-independent approach and sequence a metagenome. You get some genome bins, but you are unsure, whether they represent the species you are interested in, because they lack the 16S rRNA gene sequence as the link to your amplicon data.
This was the situation we found ourselves in when we began our project. In our research group we are studying the succession of bacteria thriving during and after phytoplankton blooms in the North Sea. We extensively use fluorescence in situ hybridization (FISH) as it allows for counting the relative abundances of bacterial clades based on 16S rRNA sequences and using FISH, we recurrently detected a flavobacterial clade named “Vis6”, which resisted laboratory cultivation. Having the 16S rRNA based probe at hand and the flow cytometer with cell sorter down the corridor, we thought about combining these two methods for a targeted genomics approach. The pipeline we had in mind introduces a 16S rRNA based FISH signal into the cells of a species of interest within a bacterial community. These cells are then sorted based on that FISH signal by fluorescence activated cell sorting (FACS), resulting in an enrichment of very limited diversity (ideally on the species level) for subsequent genomic sequencing. Genome annotation then provides clues about the capabilities and functions of the targeted species in the given environment.
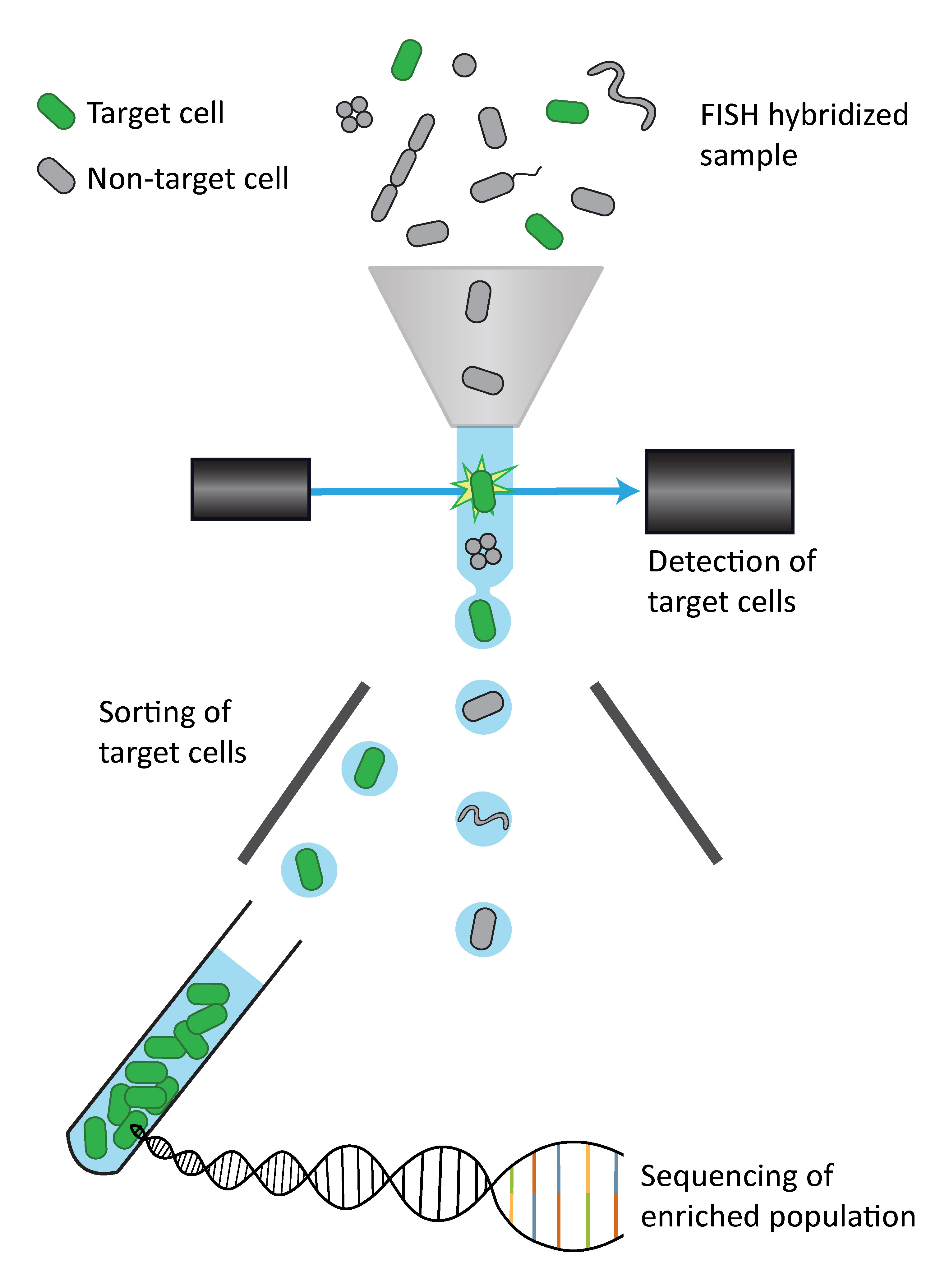
So far, so good, but we were facing two major challenges: 1.) A very bright FISH signal was needed for the detection and sorting of targeted cells using flow cytometry and 2.) Sufficient unimpaired DNA material was required for high quality genome sequencing. Our lab (Max Planck Institute for Marine Microbiology) had its expertise in FISH methods and we teamed up with DOE’s Joint Genome Institute (JGI) who brought along the sequencing expertise, in particular from little starting material. Our work consisted of a lot of troubleshooting using laboratory cultures, but eventually we developed a FISH&FACS protocol that provided high FISH signal intensities without impairing the sequencing quality by testing and validating the most suitable cell fixation approach and assessing the optimal number of sorted cells.
With this optimized pipeline at hand, we sampled seawater from the North Sea and hybridized it with our Vis6-specific FISH probe. The sorted cell enrichments were genome sequenced and the resulting mini-metagenomes of reduced diversity yielded good quality metagenome assembled genomes (MAGs). Due to the encoded 16S rRNA gene sequences and further supported by the FISH probes used, we were able to assign the taxonomic identity to the MAGs. Gene annotations of these MAGs revealed that Vis6 is a putative polysaccharide and protein degrader.
We see the main application of our pipeline in filling a gap that metagenomics and single cell genomics leave in the field of culture independent species descriptions and a valuable addition to the toolkit of cultivation-independent genomics. Metagenomes are challenged by highly complex samples and the bins are often lacking the 16S rRNA genes as a taxonomic marker. While single cell genomes most often have this taxonomic link, they are generally hampered by low genome completeness. Both methods retrieve highly abundant species more often than species of lower abundance. With a targeted genomic approach, such as the FISH&FACS pipeline we describe in our paper, specific groups or species can be genomically interrogated, opening a window into the rare biosphere, even of highly complex samples.
Follow the Topic
-
Microbiome

This journal hopes to integrate researchers with common scientific objectives across a broad cross-section of sub-disciplines within microbial ecology. It covers studies of microbiomes colonizing humans, animals, plants or the environment, both built and natural or manipulated, as in agriculture.
Related Collections
With Collections, you can get published faster and increase your visibility.
Harnessing plant microbiomes to improve performance and mechanistic understanding
This is a Cross-Journal Collection with Microbiome, Environmental Microbiome, npj Science of Plants, and npj Biofilms and Microbiomes. Please click here to see the collection page for npj Science of Plants and npj Biofilms and Microbiomes.
Modern agriculture needs to sustainably increase crop productivity while preserving ecosystem health. As soil degradation, climate variability, and diminishing input efficiency continue to threaten agricultural outputs, there is a pressing need to enhance plant performance through ecologically-sound strategies. In this context, plant-associated microbiomes represent a powerful, yet underexploited, resource to improve plant vigor, nutrient acquisition, stress resilience, and overall productivity.
The plant microbiome—comprising bacteria, fungi, and other microorganisms inhabiting the rhizosphere, endosphere, and phyllosphere—plays a fundamental role in shaping plant physiology and development. Increasing evidence demonstrates that beneficial microbes mediate key processes such as nutrient solubilization and uptake, hormonal regulation, photosynthetic efficiency, and systemic resistance to (a)biotic stresses. However, to fully harness these capabilities, a mechanistic understanding of the molecular dialogues and functional traits underpinning plant-microbe interactions is essential.
Recent advances in multi-omics technologies, synthetic biology, and high-throughput functional screening have accelerated our ability to dissect these interactions at molecular, cellular, and system levels. Yet, significant challenges remain in translating these mechanistic insights into robust microbiome-based applications for agriculture. Core knowledge gaps include identifying microbial functions that are conserved across environments and hosts, understanding the signaling networks and metabolic exchanges between partners, and predicting microbiome assembly and stability under field conditions.
This Research Topic welcomes Original Research, Reviews, Perspectives, and Meta-analyses that delve into the functional and mechanistic basis of plant-microbiome interactions. We are particularly interested in contributions that integrate molecular microbiology, systems biology, plant physiology, and computational modeling to unravel the mechanisms by which microbial communities enhance plant performance and/or mechanisms employed by plant hosts to assemble beneficial microbiomes. Studies ranging from controlled experimental systems to applied field trials are encouraged, especially those aiming to bridge the gap between fundamental understanding and translational outcomes such as microbial consortia, engineered strains, or microbiome-informed management practices.
Ultimately, this collection aims to advance our ability to rationally design and apply microbiome-based strategies by deepening our mechanistic insight into how plants select beneficial microbiomes and in turn how microbes shape plant health and productivity.
This collection is open for submissions from all authors on the condition that the manuscript falls within both the scope of the collection and the journal it is submitted to.
All submissions in this collection undergo the relevant journal’s standard peer review process. Similarly, all manuscripts authored by a Guest Editor(s) will be handled by the Editor-in-Chief of the relevant journal. As an open access publication, participating journals levy an article processing fee (Microbiome, Environmental Microbiome). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief of the journal where the article is being submitted.
Collection policies for Microbiome and Environmental Microbiome:
Please refer to this page. Please only submit to one journal, but note authors have the option to transfer to another participating journal following the editors’ recommendation.
Collection policies for npj Science of Plants and npj Biofilms and Microbiomes:
Please refer to npj's Collection policies page for full details.
Publishing Model: Open Access
Deadline: Jun 01, 2026
Microbiome and Reproductive Health
Microbiome is calling for submissions to our Collection on Microbiome and Reproductive Health.
Our understanding of the intricate relationship between the microbiome and reproductive health holds profound translational implications for fertility, pregnancy, and reproductive disorders. To truly advance this field, it is essential to move beyond descriptive and associative studies and focus on mechanistic research that uncovers the functional underpinnings of the host–microbiome interface. Such studies can reveal how microbial communities influence reproductive physiology, including hormonal regulation, immune responses, and overall reproductive health.
Recent advances have highlighted the role of specific bacterial populations in both male and female fertility, as well as their impact on pregnancy outcomes. For example, the vaginal microbiome has been linked to preterm birth, while emerging evidence suggests that gut microbiota may modulate reproductive hormone levels. These insights underscore the need for research that explores how and why these microbial influences occur.
Looking ahead, the potential for breakthroughs is immense. Mechanistic studies have the power to drive the development of microbiome-based therapies that address infertility, improve pregnancy outcomes, and reduce the risk of reproductive diseases. Incorporating microbiome analysis into reproductive health assessments could transform clinical practice and, by deepening our understanding of host–microbiome mechanisms, lay the groundwork for personalized medicine in gynecology and obstetrics.
We invite researchers to contribute to this Special Collection on Microbiome and Reproductive Health. Submissions should emphasize functional and mechanistic insights into the host–microbiome relationship. Topics of interest include, but are not limited to:
- Microbiome and infertility
- Vaginal microbiome and pregnancy outcomes
- Gut microbiota and reproductive hormones
- Microbial influences on menstrual health
- Live biotherapeutics and reproductive health interventions
- Microbiome alterations as drivers of reproductive disorders
- Environmental factors shaping the microbiome
- Intergenerational microbiome transmission
This Collection supports and amplifies research related to SDG 3, Good Health and Well-Being.
All submissions in this collection undergo the journal’s standard peer review process. As an open access publication, this journal levies an article processing fee (details here). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief.
Publishing Model: Open Access
Deadline: Jun 16, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in