
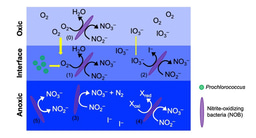
I would never expect myself to be so bothered by my success in obtaining data from experiments. What I measured was nitrite oxidation rate in samples collected from anoxic seawaters (Sun et al., 2017). Nitrite oxidation was thought to be restricted in oxic conditions and all known marine nitrite oxidizers were obligate aerobes. The detection of this microbially mediated reaction in anoxic seawaters means something weird was going on.
Very soon I found out that nitrite oxidation had been first reported from those anoxic seawaters in 1990 (Lipschultz et al., 1990), and one of the authors was my advisor, Professor Bess B. Ward, who had been bothered by this weird phenomenon much longer than me, but was constrained by techniques at that time.

Professor Bess B. Ward performing stable isotope incubation experiment to study various nitrogen cycling processes in anoxic seawaters onboard R/V Nathaniel B. Palmer in 2013. Samples in (Sun et al., 2019) were collected by Dr. Amal Jayakumar on this cruise

Xin Sun on the R/V Sally Ride after sampling thousands of bottles of seawater to study nitrite oxidation and nitrite oxidizers in 2018, and Professor Bess B. Ward was the chief scientist
The mystery of nitrite oxidation in anoxic seawaters kept me awake at night. In the winter of 2016, an idea came to us: the fact that we could detect the reaction regardless of oxygen means that nitrite oxidizing bacteria must be present in situ in those anoxic seawaters! If they were those normal nitrite oxidizers breathing oxygen, they would not choose to live there. I got so excited about this thought and was eager to find out who these weirdos are. The idea of using metagenomics came to my mind. I talked to Bess about metagenomics when I had no experience in either assembling any genome or running any code in Linux. She not only said a solid yes to me with funding support but also introduced me to Dr. Maggie CY Lau’s metagenomic lab lecture as part of Professor Tullis C. Onstott’s Geomicrobiology course and a year later, introduced me to Dr. Sebastian Lücker from Radboud University whose lab has a metagenomic pipeline, which improved the quality of the genomes. Then collaboration thrived. And we did find the metagenomes of the weird nitrite oxidizers in anoxic seawater!
I can hardly remember how many exciting and nervous moments happened in Bess’s office: two highlights were the successful assembly of two draft genomes of nitrite oxidizers from anoxic seawaters and the publication of draft genomes of nitrite oxidizers from mainly oxic seawaters (Pachiadaki et al., 2017) around the time when we submitted our paper to ISMEJ (Sun et al., 2019). I don’t know how many times I emailed Maggie about coding errors or how many WhatsApp messages Linnea FM Kop in the Lücker lab and I sent to each other about metagenomic analyses. I do remember how enthusiastic everyone was and the details of all the reviewers’ comments, which made the paper better. Finding a counterintuitive outlier indeed could be the start of an amazing research project, and microbes are so good at being outliers.
References
Lipschultz F, Wofsy SC, Ward BB, Codispoti LA, Friedrich G, Elkins JW. (1990). Bacterial transformations of inorganic nitrogen in the oxygen-deficient waters of the Eastern Tropical South Pacific Ocean. Deep Sea Res Part A, Oceanogr Res Pap 37: 1513–1541.
Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK, et al. (2017). Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science 358: 1046–1051.
Sun X, Ji Q, Jayakumar A, Ward BB. (2017). Dependence of nitrite oxidation on nitrite and oxygen in low oxygen seawater. Geophys Res Lett 44: 7883–7891.
Sun X, Kop LFM, Lau MCY, Frank J, Jayakumar A, Lücker S, et al. (2019). Uncultured Nitrospina-like species are major nitrite oxidizing bacteria in oxygen minimum zones. ISME J.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in