Fun-sized Fungal Genomics: Expanding biological knowledge with single cells
Published in Microbiology
[Our paper in Nature Microbiology can be found here.]
Various estimates suggest there’s anywhere from 1 million to 5 million different fungal species, only 10% of which have been described. Furthermore, much of what we as a scientific community know about fungi is limited to those species that are model genetic systems, or industrially or medically important. The vast majority of these fungi predominantly reside in two fungal phyla, collectively known as the Dikarya and comprising the Ascomycota and Basidiomycota. However, current evolutionary studies1 demonstrate that the fungal tree stretches back about a billion years and is populated by seven diverse fungal phyla, including the hyper-diverse but vastly understudied Cryptomycota2.
Clearly we’re missing something big.
Most of what’s missing belongs to species that resist culturing. Environmental rDNA surveys predict their presence, but they don’t show up in plate-based screens and are inaccessible to genome sequencing. This is the so-called “fungal dark matter,” and a better understanding of their genomes will provide clues regarding not only evolutionary history, but also basic biology and metabolism.
That’s where single-cell genomics comes in. Single-cell genomics methods rely on isolation and lysis of individual cells with subsequent whole genome amplification and sequencing. While the application of these methods has sharply increased in recent years, most of the current environmental work has focused primarily on bacterial and archaeal systems.
The US Department of Energy (DOE) Joint Genome Institute (JGI) collaborates with scientists around the world to answer large-scale or otherwise challenging biological questions related to energy and the environment. One scientist, Dr. Timothy James at the University of Michigan, wanted to understand the biology and evolutionary history of unknown and unculturable environmental fungi.
To do so, we started by developing methods for robust capture and de novo assembly of environmental fungal genomes, and then applied these methods for the first time to fungi which are evolutionarily diverse but similar in their resistance to traditional culturing efforts: Rozella allomycis, Caulochytrium protostelioides, Dimargaris cristalligena, Piptocephalis cylindrospora, Syncephalis pseudoplumigaleata, and Thamnocephalis sphaerospora. We focused primarily on biotrophic mycoparasites because they are an understudied group of fungi, but also included the pollen saprotroph Blyttiomyces helicus and Daphnia parasite Metschnikowia bicuspidata.
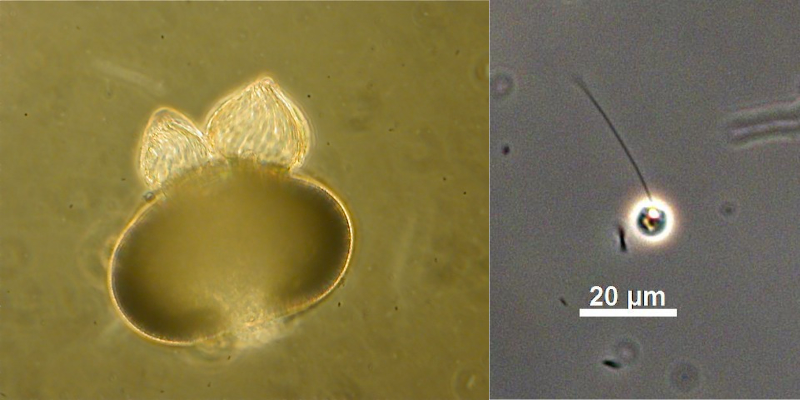
Many people were involved in this effort, which spanned over 5 years. From sampling to sequencing, assembly to annotation, groups within and outside of JGI helped bring this project together. The project had already been ongoing for a few years when I came onboard in 2015 as a UC Berkeley postdoc with Dr. Igor Grigoriev, head of the JGI's Fungal Program and an adjunct professor at UC Berkeley. Dr. Alisha Quandt, now an Assistant Professor at the University of Colorado Boulder, provided invaluable analyses for this project as a postdoc with Tim. JGI Senior Research Associate Dr. Doina Ciobanu worked to tackle the benchmarking techniques necessary to translate the JGI’s existing microbial single-cell workflow to fungi, which introduce more challenges including cell wall, ploidy, and wide GC variation.
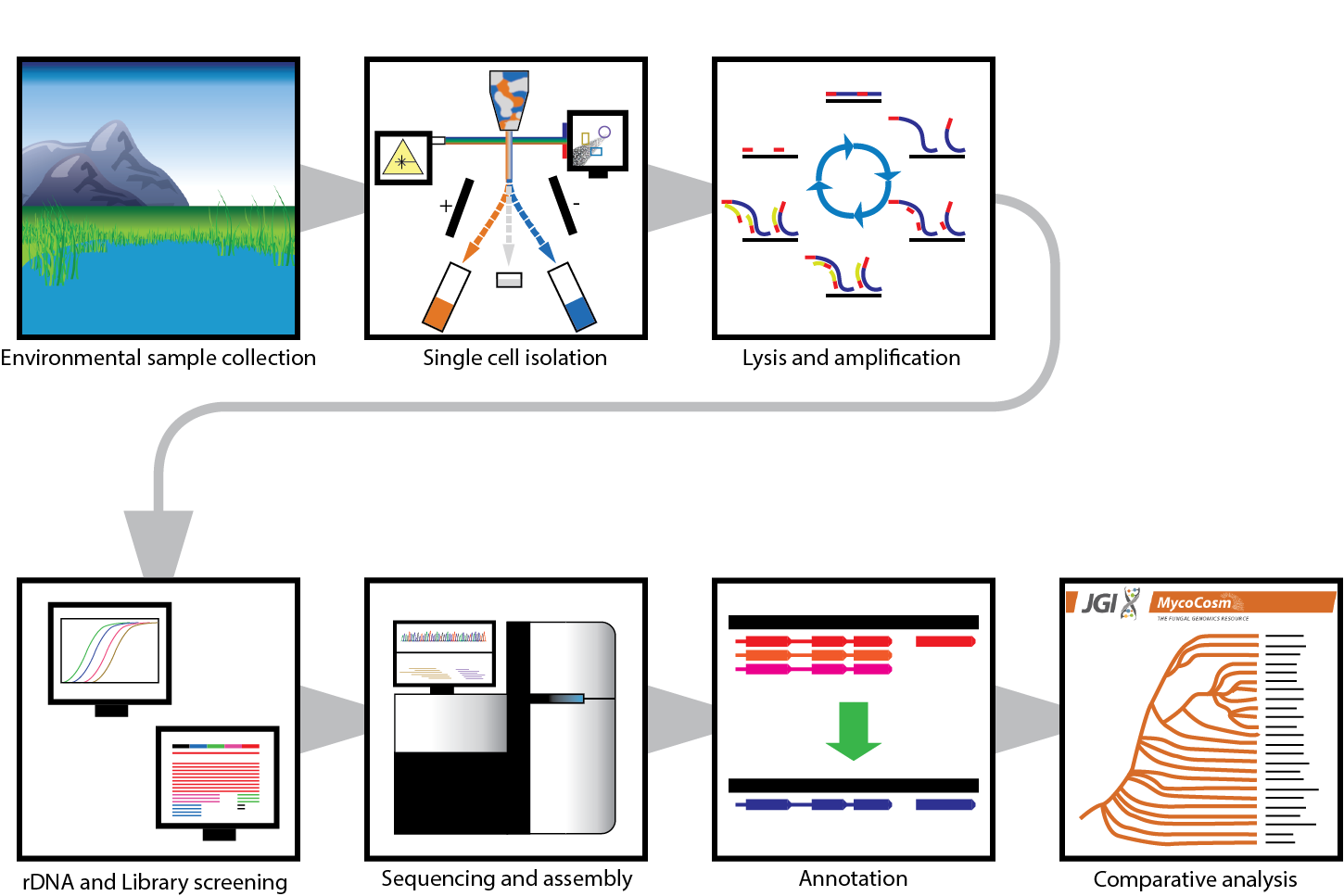
Our pipeline provided new genomes for understudied clades, with which we explored the biology and evolutionary histories of these uncultivated species. But the really exciting thing with this work is where we go next. In the near future, this approach will be adapted to sequence 50 unknown early-diverging fungi from aquatic environments. The phylogenetic relationships among the basal lineages of the fungal tree of life are the least resolved. Understanding these relationships will be crucial to having a more complete fungal tree of life, and single-cell genomics methods are key to that understanding.
The JGI news release for this publication can be found here.
References
- Spatafora et al. 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia, 108, 1028-1046. doi:10.3852/16-042
- Jones et al. 2011. Discovery of novel intermediate forms redefines the fungal tree of life. Nature, 474, 200-203. doi:10.1038/nature09984





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in