
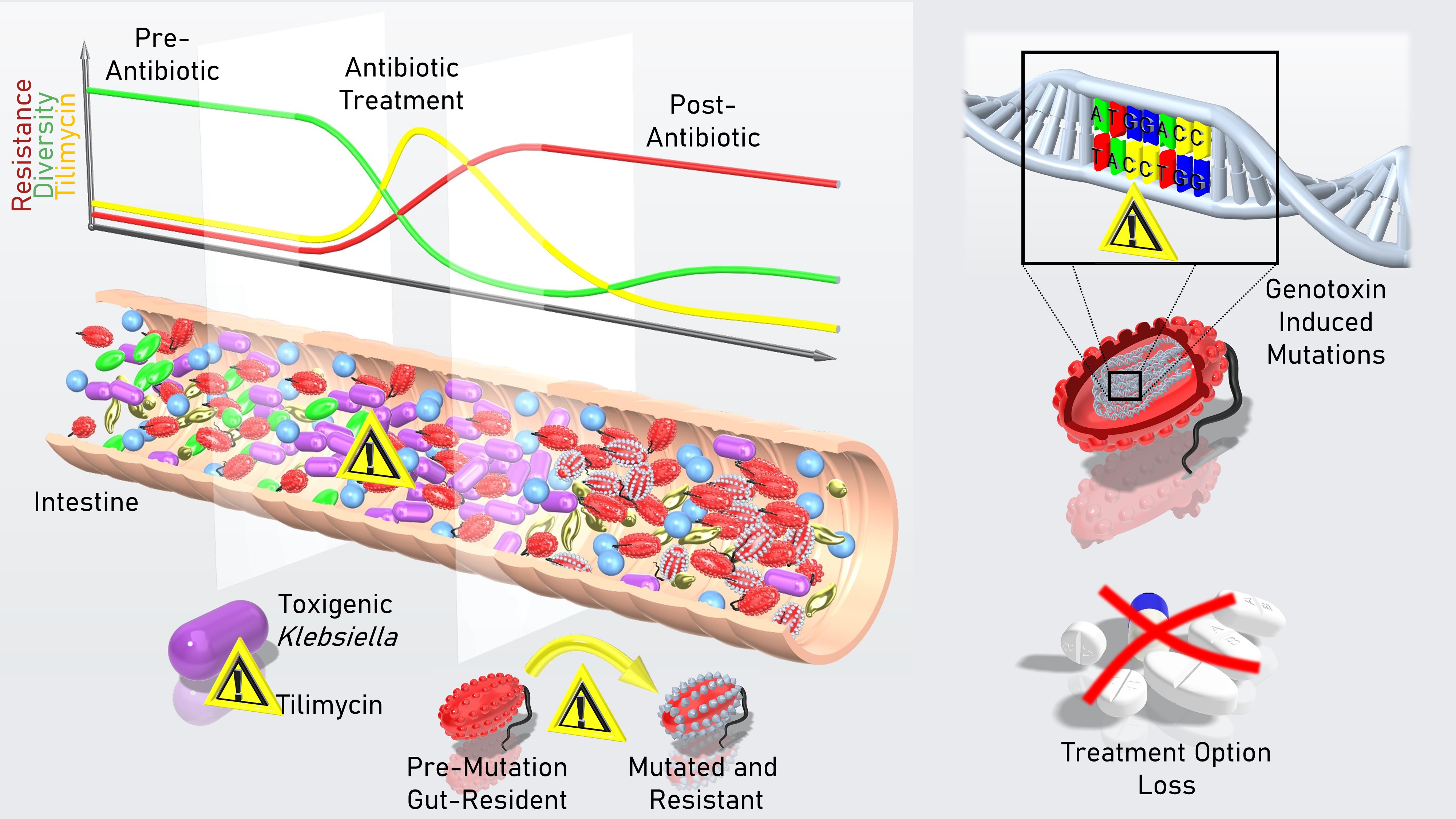
Graphical Summary. During antibiotic treatment, numbers of toxigenic Klebsiella in the gut surge and the genotoxic metabolite they produce, tilimycin, accumulates in the intestine. Tilimycin damages bacterial DNA, creating mutations and killing multiple species. Loss of microbial diversity and mutational pressure alters evolution of the community. Surviving bacteria carry mutations including new resistance to antibiotics. As a consequence, subsequent antibiotic treatment can foster multi-resistant populations. Picture and Video (below) credit: F. Feist
Context
Our research is motivated by the question of how bacteria interact with each other and their host in the densely populated ecosystem of the human gut. The intestine is host to an amazing diversity of microorganisms and the tens of thousands of natural products that they secrete1. Research efforts are providing increasing clarity concerning the roles of specific microbiota-derived molecules as regulators of interactions between microbes or host cellular responses that either contribute to human health or disease. Yet, discerning the functions of specific effectors in this rich chemical cocktail is challenging and adequate model systems are often lacking.
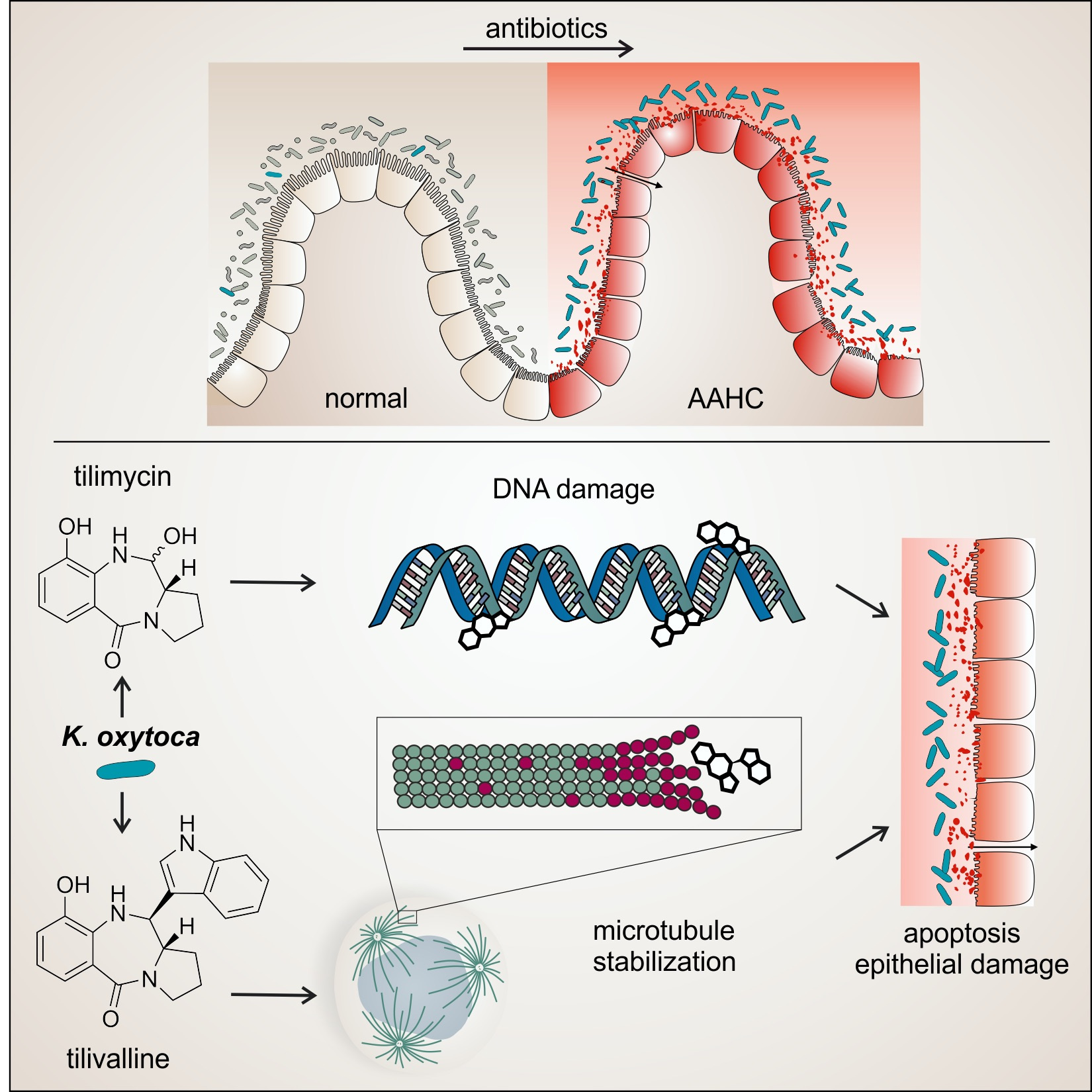
Members of the human gut microbiota include some toxigenic bacteria, such as Klebsiella species. We and others have sought to understand how these organisms and their cytotoxic products interact with the intestinal epithelium to cause acute intestinal disease. In early studies it was established that gut resident K. oxytoca is the causative agent of antibiotic-associated hemorrhagic colitis (AAHC)2,3 and that the two secreted enterotoxins, tilimycin (TM) and tilivalline (TV) damage epithelial cells in vivo3,4. Moreover, the chemical nature and the biosynthesis pathway behind the small-molecule toxins were resolved5–7 and analytical tools to quantify the toxins present in the body during disease were developed8.
More recently, the biosynthesis genes (til) for the nonribosomal peptides TM and TV were found to be conserved in related members of the human gut microbiota, K. grimontii and K. michiganensis9–11. Unterhauser et al.4 showed that disease-causing damage to the intestinal epithelium occurs when TV binds and stabilizes microtubules in a polymerized state inducing mitotic arrest and apoptosis4. By contrast, TM directly alkylates DNA bases resulting in genetic instability and genome fragmentation4,7. DNA damage caused by an alkylating agent such as TM need not be lethal, but can be repaired by error prone polymerases that create mutations. Even small doses of a genotoxin can drive genetic variation in surviving cells.
Figure 1. During antibiotic use, the number of toxigenic K. oxytoca increases dramatically. Secreted enterotoxins Tilimycin (TM) and Tilivalline (TV) bind and damage cellular targets causing pathological alteration of the intestinal lining. Picture credit: L. Pöltl
The Paper
DNA is clearly a cellular target that is shared by humans and bacteria alike. Our paper in Nature Microbiology illustrates the scope of functional consequences that can emerge when Klebsiella spp carry the til genes. We asked how the release of a stable, diffusible genotoxin impacts the gut microbiota itself. Particularly we were interested to learn how broadly effective TM might be in killing bacterial competitors in the gut ecosystem and whether the mutations created in surviving gut residents could promote clinically relevant outcomes for human carriers.
Toxigenic Klebsiella spp colonize the human intestine within days after birth and can persist in this ecosystem over a person’s lifetime. The size of the toxigenic population and the amount of til enterotoxins that accumulate in vivo can increase dramatically when the gut ecosystem is destabilized, for example, during antibiotic use. Erosion of the intestinal epithelium and colitis is one acute outcome of an intestinal bloom of til bacteria in infants, children, and adults. But what are the consequences for the resident microbiota?
We show that even low amounts of TM can inhibit the growth of several bacterial phyla present in human stool. In mice we show that TM secretion is a highly effective means for toxigenic Klebsiella spp to antagonize competitors. TM accumulation in the murine intestine exerted cross-phyla antibiotic activity against a complex flora. Presence of the genotoxin exacerbated depletion of the gut microbiota during amoxicillin treatment, reduced richness and altered the taxonomic composition of the succession community following exposure. In addition to antibiotic mortality, the hypermutator environment created by TM drove genetic variation in the surviving bacterial population.
Like other members of the pyrrolobenzodiazepine substance class, TM prefers purine substrates in a non-specific sequence context, which is compatible with genome-wide activity. Accordingly, the functional consequences were predicted to be broad. As an example of clinically relevant mutational gain of function we demonstrated the de novo emergence of antibiotic resistance in colonized mice. The mutagenic environment created by a single asymptomatic event of genotoxic Klebsiella outgrowth was sufficient to expand the array of antibiotic resistance determinants available to gut bacteria and co-resident opportunistic pathogens such as Klebsiella pneumoniae and Escherichia coli.
We conclude that TM is a bacterial mutagen, and flares of genotoxic Klebsiella have the potential to drive the emergence of resistance, destabilize the gut microbiota and shape its evolutionary trajectory.
Video credit: F. Feist
(Video edited - Version 1.2 )
Open Questions
These findings in mice raise several important questions for human carriers of genotoxic bacteria. As TM is active in both human and bacterial cells, we speculate that off-target effects of antibiotic use puts colonized patients at risk for species depletion, antibiotic-associated diarrhea, and intestinal disease. It is further possible that TM-mediated killing of bacteria, and subsequent cell lysis would release immunostimulatory compounds such as lipopolysaccharides. This in turn may amplify inflammation in addition to direct toxicity to human cells. We predict that genotoxic Klebsiella spp. in human carriers also drive mutational evolution of gut bacteria and thereby expand the reservoir of resistance genes. Could that put individuals at risk for future resistant infections originating from the commensals and opportunistic pathogens they carry?
Our findings may be particularly relevant in infancy. The microbial community in the infant gut is dynamic during the first years of life12–16. Maturation of these early communities contributes importantly to a child’s metabolic and immune development17–19. Our study thus raises concern that early life experience with antibiotics, medications or dietary changes that trigger dysbiotic outgrowth will add the additional pressure of TM exposure in the infant gut. In such cases increased microbial volatility and concomitant loss of community resilience could further impair this critical phase of microbial maturation, potentially shift its evolutionary trajectory and predispose these children to immune-mediated disorders later in life18–20.
References
- Shine, E. E. & Crawford, J. M. Molecules from the Microbiome. Annu. Rev. Biochem. 90, 789–815 (2021).
- Högenauer, C. et al. Klebsiella oxytoca as a Causative Organism of Antibiotic-Associated Hemorrhagic Colitis. N. Engl. J. Med. 355, 2418–2426 (2006).
- Schneditz, G. et al. Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. Proc. Natl. Acad. Sci. USA 111, 13181–13186 (2014).
- Unterhauser, K. et al. Klebsiella oxytoca enterotoxins tilimycin and tilivalline have distinct host DNA-damaging and microtubule-stabilizing activities. Proc. Natl. Acad. Sci. USA 116, 3774–3783 (2019).
- Dornisch, E. et al. Biosynthesis of the Enterotoxic Pyrrolobenzodiazepine Natural Product Tilivalline. Angew. Chem. Int. Ed. 56, 14753–14757 (2017).
- Tse, H. et al. A tricyclic pyrrolobenzodiazepine produced by Klebsiella oxytoca is associated with cytotoxicity in antibiotic-associated hemorrhagic colitis. J. Biol. Chem. 292, 19503–19520 (2017).
- Alexander, E. M. et al. Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic Pathogen Klebsiella oxytoca. ACS Infect. Dis. 6, 1976–1997 (2020).
- Glabonjat, R. A. et al. Simultaneous quantification of enterotoxins tilimycin and tilivalline in biological matrices using HPLC high resolution ESMS2 based on isotopically 15N-labeled internal standards. Talanta 222, 121677 (2021).
- Shibu, P. et al. Improved molecular characterization of the Klebsiella oxytoca complex reveals the prevalence of the kleboxymycin biosynthetic gene cluster. Microb. Genom. 7, 000592 (2021).
- Leitner, E. et al. Improved diagnosis of antibiotic-associated haemorrhagic colitis (AAHC) in faecal specimens by a new qualitative real-time PCR assay detecting relevant toxin genes of Klebsiella oxytoca sensu lato. Clinical Microbiology and Infection 28, S1198743X21005371 (2022).
- Cosic, A. et al. Variation in accessory aenes within the Klebsiella oxytoca species complex delineates monophyletic members and simplifies coherent genotyping. Front. Microbiol. 12, 692453 (2021).
- Bäckhed, F. et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 17, 690–703 (2015).
- Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
- Subramanian, S. et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421 (2014).
- Bokulich, N. A. et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8, (343):343ra82 (2016).
- Yassour, M. et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, (343):343ra81 (2016).
- Vatanen, T. et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 (2018).
- Vatanen, T. et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 842–853 (2016).
- Olin, A. et al. Stereotypic Immune System Development in Newborn Children. Cell 174, 1277-1292.e14 (2018).
- Stokholm, J. et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun 9, 141 (2018).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in