Genes, Milk, and Nitrogen: Decoding How Cows Manage Waste at the Genomic Level
Published in Genetics & Genomics, Agricultural & Food Science, and Zoology & Veterinary Science
Explore the Research
 Research Communities by Springer Nature
Research Communities by Springer Nature
What do we really know about dairy and chronic diseases?
Dairy fats, inflammation and the food matrix
Published in Dairy Science and Management
October 2025
The Study at a Glance
Researchers analyzed more than 347,000 milk test-day records from 52,000 Iranian Holstein cows, using genomic data from 2,187 bulls and over 41,000 SNP markers.
By applying a single-step genome-wide association study (ssGWAS) combined with random regression models, they tracked how genetic effects on MUN changed over time across the lactation cycle—revealing the dynamic nature of nitrogen metabolism in dairy cows.
What They Found
-
Genetic control of MUN varies by lactation stage—only three SNPs overlapped between early, mid, and late lactation.
-
Seventeen candidate genes were identified, including PGLYRP3, S100A9, ABCB11, CYP1A1, and SLC5A11, linked to nitrogen metabolism, immune response, and nutrient transport.
-
Many of these genes lie near quantitative trait loci (QTLs) associated with milk composition, fertility, and health traits—highlighting how nitrogen efficiency connects to multiple aspects of dairy performance.
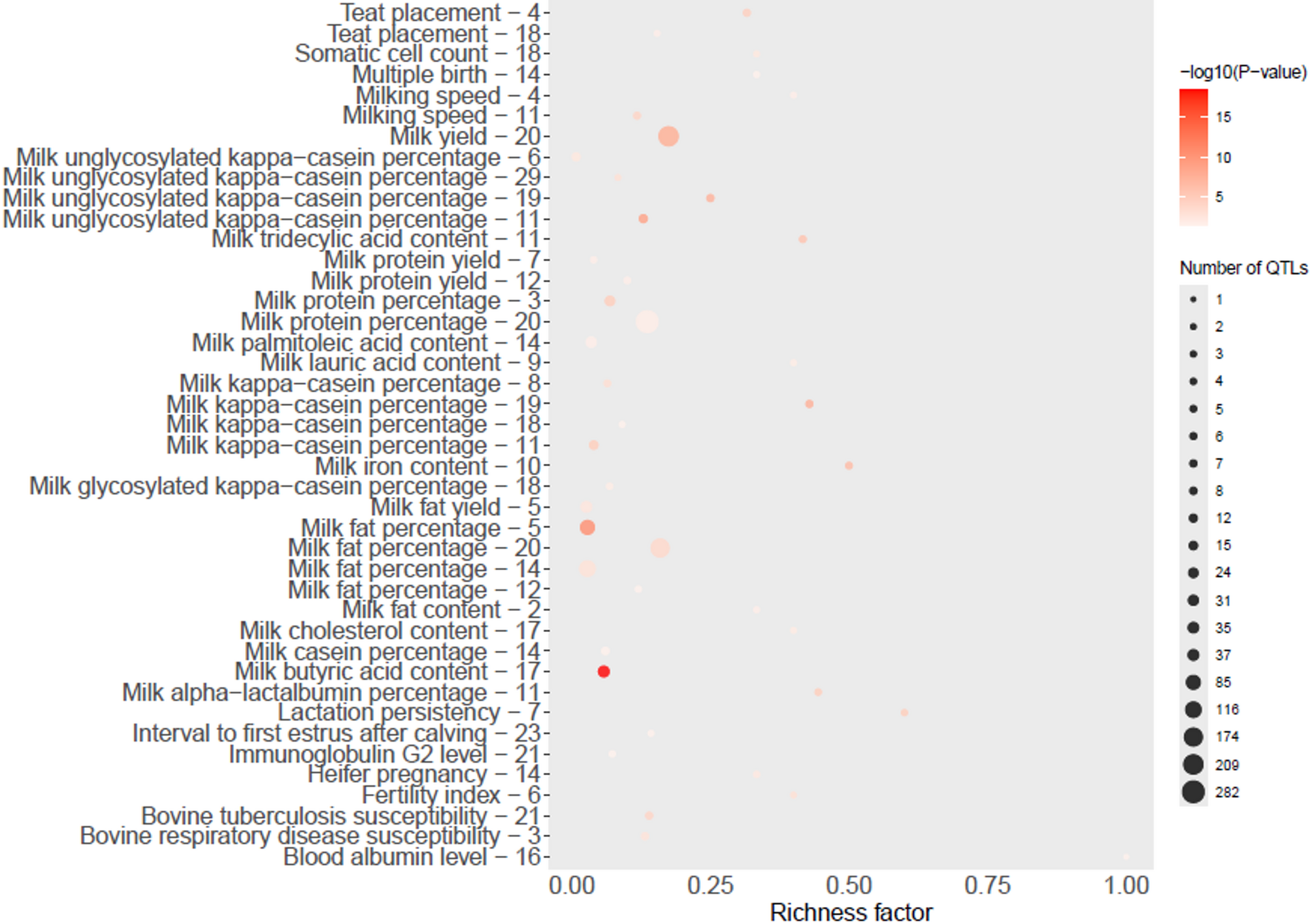
This figure shows enriched QTLs associated with MUN across lactation stages. Traits such as milk protein content, fertility, and disease resistance share common genomic regions — suggesting that nitrogen efficiency is intertwined with both productivity and animal health.
Figure 2 from Mortazavi et al. (2025), Dairy Science and Management. Shared under CC BY-NC-ND 4.0. https://doi.org/10.1186/s44363-025-00015-9
Why It Matters
Understanding the genetic architecture of MUN can help breeders select cows that use nitrogen more efficiently, reducing nitrogen losses and improving sustainability. This approach can help cut emissions from manure and urine without sacrificing milk yield or fertility — a major step toward climate-smart dairy production.
Looking Ahead
Mortazavi et al. recommend that future research should:
-
Validate the identified candidate genes through functional and expression studies
-
Explore genotype–environment interactions to assess how management and diet influence nitrogen efficiency
-
Incorporate MUN into multi-trait genomic selection programs to balance production, fertility, and sustainability
Full Paper
Mortazavi, M., Zandi, M.B., Pahlavan, R., Eskandari Nasab, M., Mulim, H.A., & Rojas de Oliveira, H. Genome-wide association analysis based on random regression models for milk urea nitrogen in Iranian Holstein cattle. Dairy Sci. Manag. 2, 12 (2025). https://doi.org/10.1186/s44363-025-00015-9
Follow the Topic
-
Dairy Science and Management

This is an open access peer-reviewed journal that aims to publish innovative research about the management of dairy animals, the production of dairy products and the related food security considerations.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in