Getting a handle on the evolutionary processes of cancer
Published in Ecology & Evolution
The paper in Nature Ecology & Evolution is here: https://go.nature.com/2N9UD1S
In response to the massive decline in sequencing costs, the tried and tested single-biopsy, few-loci experimental strategy has recently yielded to “multi-biopsy sampling” and next generation sequencing. It seems to me that 21st century cancer genetics is now very much about casting the experimental net wide and using mathematical modelling to reveal novel evolutionary information. Measuring intra-tumour heterogeneity is particularly key since it permits phylogenetics and other analyses to be employed with the potential to discover how precisely cancers develop.
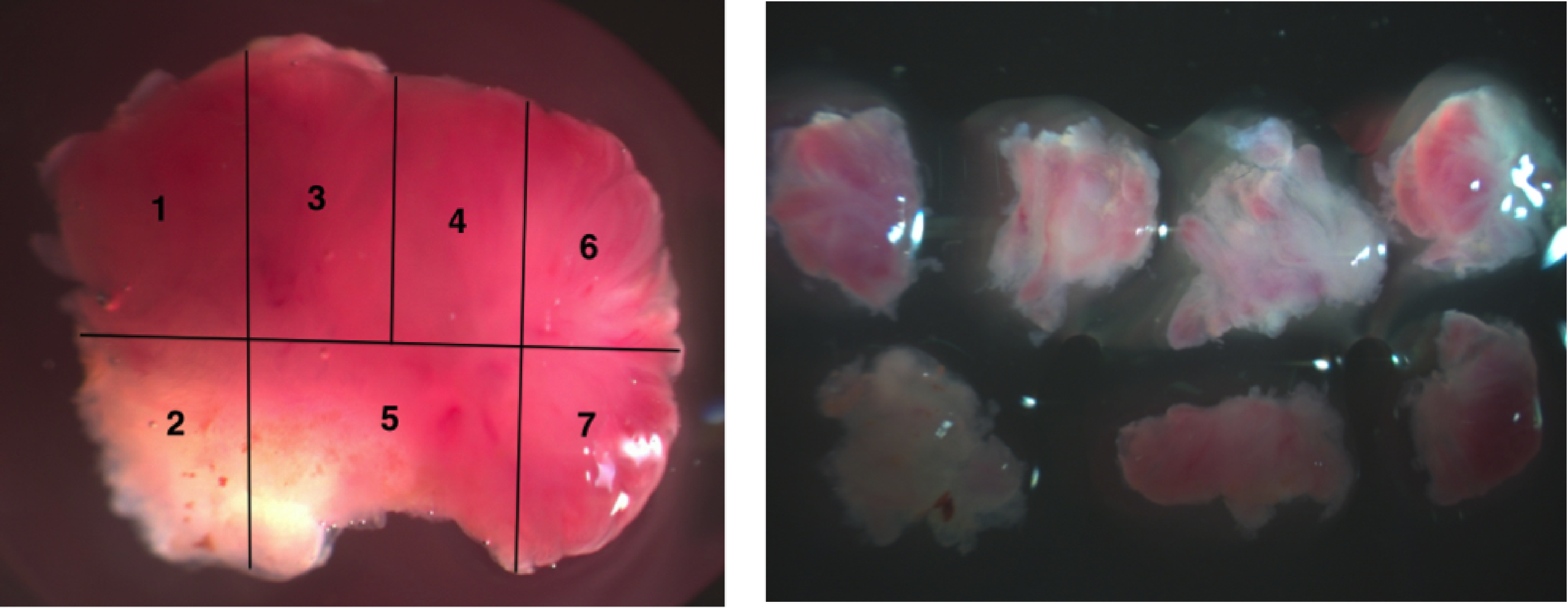
Sampling tumours spatially is essential to perform evolutionary analyses. Micro-dissecting small lesions, such as adenomas, presents an extra challenge.
Colorectal cancer is a well studied malignancy and a special interest to myself and our group. It is true that many of the common genetic features (so-called "driver mutations") are actually already known, but then again, many challenging questions remain unanswered. For example, what is the precise relationship between genotypes and cancer phenotypes? This is to suggest that we simply don't know which of the key mutations are responsible for the transformation of benign tissues to cancers.
The motivations for this study were therefore clear: we wanted to improve the understanding of colon cancer genetics and in particular how adenomas (which are benign precursor lesions) evolve to become cancers. However, in order to do this we needed to overcome certain practical problems. Adenomas, you see, are common and routinely removed during endoscope procedures in the clinic. However, to acquire fresh frozen samples (and therefore high quality sequence data) a special process needed to be in place to secure specimens as they were removed. Furthermore, as I have mentioned above, multi-region sampling was also necessary to allow evolutionary inferences, but adenomas are usually small and hard to micro-dissect. The pathologist handling each patient also requires enough material be to set aside to perform his or her diagnostic work, which means we can not ever have free reign of the scalpel.
How did we overcome these hurdles? Among the authors are several gastroenterologists and through them we were able to acquire the correct ethics and specially arrange collections from John Radcliffe Hospital. To physically sample the adenomas, it turned our, we simply needed patience and the willing to coordinate with the on-duty pathologist. And of course a steady hand.
The most surprising finding of this study came as we build phylogenetic trees from the adenoma sequencing data. In at least three of the nine lesions sampled we found that key driver mutations coexisted across the tissues (see manuscript figure 2 and the figure below), meaning that they could not have been acquired sequentially or as selective sweeps in the way that has almost universally been assumed. Notably in one adenoma, for which we had four biopsies, one biopsy had a mutation in the KRAS gene while the other three had mutations in TP53, which is a gene commonly attributed to cancers (though this lesion was verified by two pathologists to be actually an adenoma). Cancers, by contrast, always seem to have the same driver mutations present in all biopsies.
Two models of carcinogenesis: our data suggests that not all cancers follow the classical selective sweep model
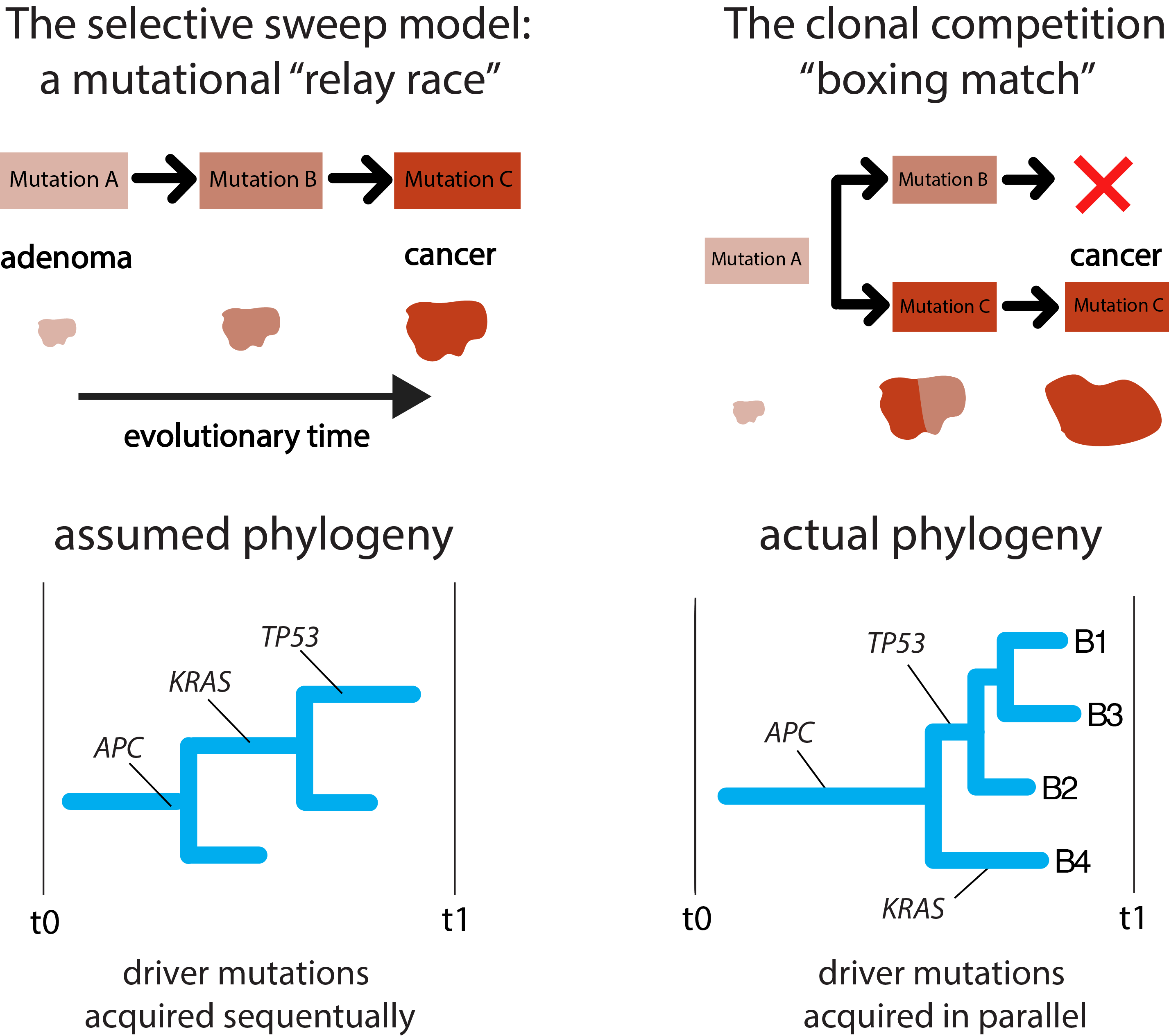
If we put the evidence together we see that the evolutionary path from normal tissue to adenoma and then cancer must be more akin to clonal competition resulting in a single sweep rather than several rounds of selective sweeps as previously assumed: colorectal cancer, it seems, is not always a driver mutation relay race, but a boxing match.
Although the classical model of colorectal cancer may remain true in some cases, in the light of evolutionary analyses it certainly seems that we need to rethink certain aspects of it.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in