How amino acid repeats are modifying our understanding of host-pathogen arms race?
Published in Ecology & Evolution, Genetics & Genomics, and Immunology
In the beginning was the Idea
During the Covid lockdown, my interest was piqued by the dynamics of host-pathogen interaction. It's truly captivating how a limited number of immune cells and pathways in the host consistently combat an overwhelming array of pathogens daily. The constant interplay between host and pathogens, each striving to outdo the other, fascinated me, leading to a coevolutionary arms race.
Drawing from existing knowledge, I knew that pathogens apply selection pressure on host immunity, prompting rapid functional diversification of immune genes—a phenomenon observed in various vertebrate clades. While delving into the literature on host-pathogen interaction mechanisms, we (Lokdeep Teekas, Sandhya Sharma, and Dr Nagarjun Vijay) stumbled upon a research article highlighting the use of Leucine-rich repeats by both parties to counteract each other.
The notion of protein repeats playing a pivotal role in immunity caught our attention. Protein repeats are renowned for their contribution to rapid evolutionary diversification, exhibiting a rate up to 100,000 times faster than point mutations. And voila! This marked the commencement of a fascinating journey filled with intriguing discoveries. Not only did these findings become the focus of numerous upcoming research articles from our lab, but they also formed the central theme of my PhD thesis.
Embarking on the Immune Odyssey: Amino Acid Repeats Unveiled
Working on immune genes and amino acid repeats provided both excitement and challenges. These topics are actively researched, covering evolutionary consequences to medical implications. Repeat length variation, illustrated in RUNX2 QA repeat in dogs, and Huntington’s disease due to polyQ expansion in HTT offers morphological diversification. Combining knowledge of repeat length variation for immune genes presented exciting challenges and required innovative approaches.
Our main challenge was empirically identifying repeat length variation within orthologous repeats across Tetrapoda's phylogeny (Figure 1). This involved identifying orthologous repeats in orthologous genes and quantifying length variation between sister clades in a phylogeny.

To achieve this, we downloaded annotated protein-coding immune genes for well-annotated Tetrapod species. Amino acid repeats, and their coordinates were identified in each gene sequence. Homologous repeats were those with overlapping coordinates between the same gene sequence across species. We calculated the length contrast between sister clades using phylogenetically independent contrasts (PIC). The sister clade with a smaller average repeat length was termed contracted against the other sister clade with a larger average length for the same homologous repeat.
One Small Variation for Repeat, One Giant Leap for Evolution
Our repeat length variation analyses across all available protein-coding genes in Tetrapoda species revealed length variation in homologous repeats between sister clades. Notably, the PolyP repeat of FASLG showed significant variation between two sister clades of Rodentia ( Figure 2: expansion in Cricetidae against Muridae). We identified promising candidate genes with drastically changed repeat lengths between species or clades, contributing to evolutionary novelty.
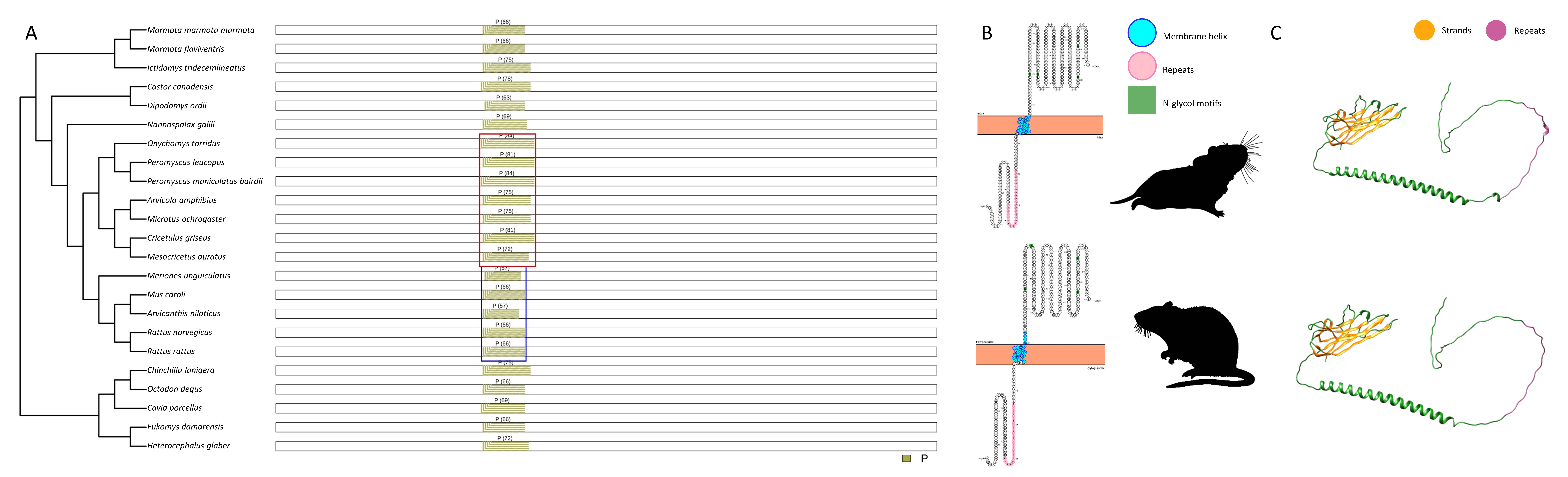
We extended the study by calculating the number of genes showing repeat expansion or contraction in each species and quantifying it across the phylogeny to identify general patterns of overall length contrast between species and species groups. Some repeats showed little to no length variation across species, possibly due to a critical length-dependent biological function. All necessary codes for result reproducibility are available on the Github repository.
Exploring the Rabbit’s Hole
After completing the project, I (Lokdeep Teekas) continued my scientific odyssey on amino acid repeats. I wanted to explore their role in an evolutionary context and disorders, so I decided to make it my PhD thesis topic. In the last year, I completed a project exploring global patterns of amino acid repeats across all available Tetrapoda species for all protein-coding genes (first author), length volatility of repeats in birds (co-author), and the role of amino acid repeats and their interactions in human disorders (first author). The pre-prints for global patterns and human disorders are available on BioRxiv for review and feedback.
Being adept at handling large-scale next-generation data using bioinformatics approaches, I hope to continue exploring different research topics in my scientific journey. May the force be with me in finding a Postdoc position to continue my journey.
Follow the Topic
-
Genes & Immunity

This journal emphasizes studies investigating how genetic, genomic and functional variations affect immune cells and the immune system, and associated processes in the regulation of health and disease.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in