How Multimodal AI Is Transforming Colorectal Cancer Diagnosis?
Published in Cancer, Computational Sciences, and General & Internal Medicine

Explore the Research
Multimodal analysis of whole slide images in colorectal cancer
npj Digital Medicine - Multimodal analysis of whole slide images in colorectal cancer
Cancer diagnosis traditionally relied on microscopical observation of tissue samples, which reveal crucial morphological details. Clinical, genomic, and biomarker data provided complementary insights that imaging alone cannot capture. Multimodal artificial intelligence (AI) models that combine whole slide images (WSI) with clinical or molecular data are becoming more common. They promise deeper insights than image-only models and can reshape how colorectal cancer (CRC) is diagnosed and understood. But how far has this field progressed? Our systematic review aimed to answer this question by examining all published studies that integrated WSI with non-image data in CRC.
.png)
Why we did it ?
With rapid surge in visual-language and multimodal models in the last few years, we were curious about how researchers are building multimodal CRC models. It was unclear which feature extraction strategies were being used for WSI, how image and non-image data were being fused, what datasets were available, and which downstream prediction tasks were being targeted. Most importantly, there is no consolidated overview of whether these approaches are consistent, comparable, or even feasible at scale. This combination of a clear knowledge gap and methodological curiosity motivated us to conduct a systematic review to understand the current state of multimodal WSI research in colorectal cancer.
To further explore these gaps and discuss our preliminary results with the community, we also conducted a workshop at MedInfo25, bringing together researchers working on digital pathology and multimodal modeling. The workshop enabled us to exchange perspectives, refine our research questions, and highlight open challenges in multimodal fusion.
What we did ?
We performed a structured systematic review following standard guidelines. We searched major scientific databases for studies that combined WSI with at least one additional data modality, such as clinical variables, genomic features, molecular markers, or other patient information. After screening titles, abstracts, and full texts, we included only studies that clearly integrated multiple data types within a unified model or analysis pipeline. We then extracted details on the methodology, datasets, multimodal fusion strategies, evaluation approaches, and reporting quality.
What we found ?
Our findings revealed that research is emerging, but still very early in its development and translation to practice. We identified 22 studies that met our criteria, a surprisingly small number given the rapid growth of AI in pathology. Most published work still relies on single-modality models, and only a limited set of studies truly integrates WSI with clinical or genomic data. We also found that TCGA cohort is a large, publicly available cancer genomics resource that includes matched whole-slide images, clinical data, and molecular profiles remains the dominant dataset, often supplemented with small private cohorts to increase the sample size.
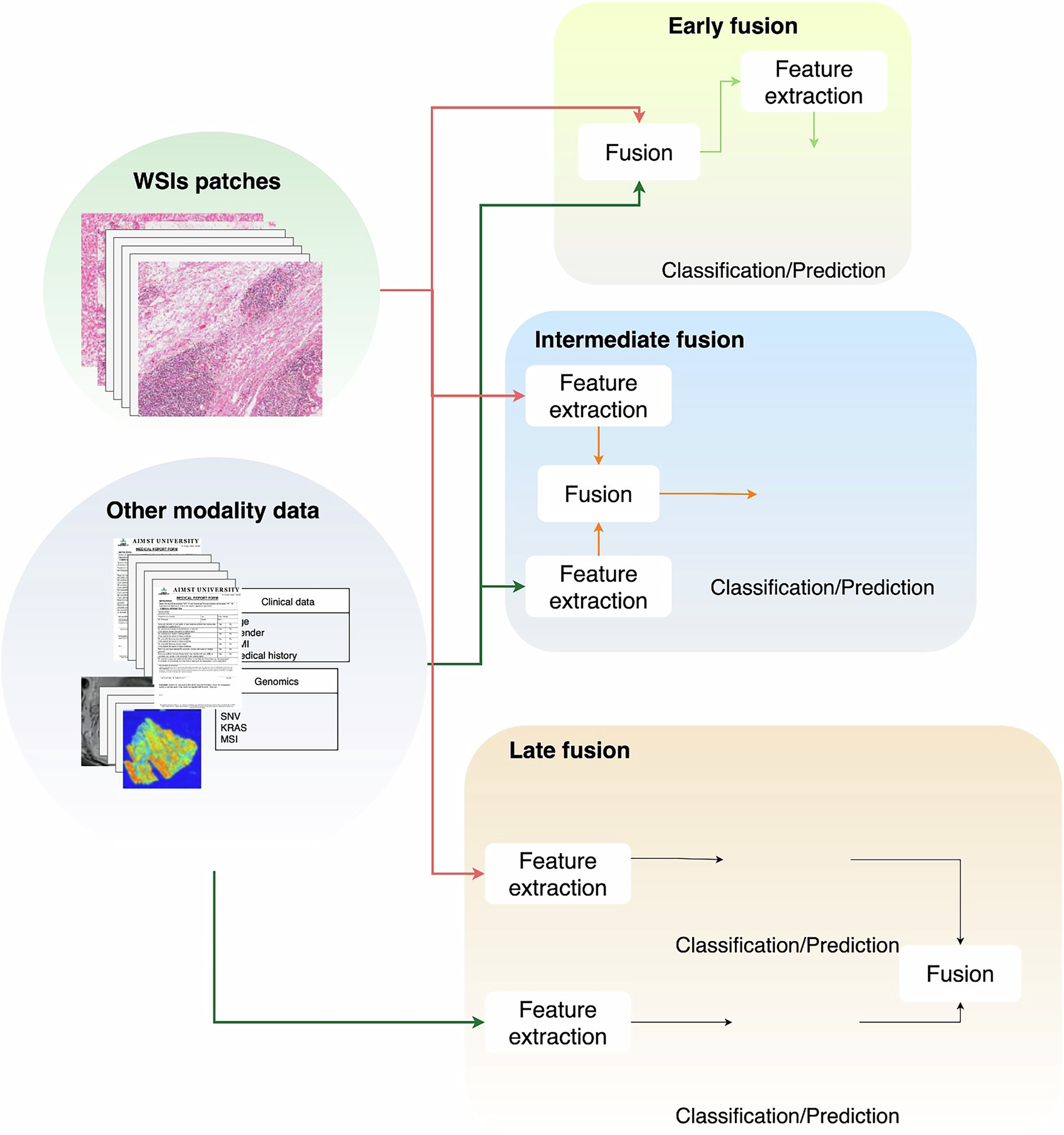
A clear pattern emerged in how the researchers processed the data. In computational pathology, feature extraction is essential because WSIs are extremely large and typically lack detailed pixel-level labels. As a result, images must be tessellated into patches, treated as a “bag of instances”, and analysed using multiple instance learning. Most studies used patch-level features extracted from ResNet-based encoders, a widely adopted deep convolutional neural network known for its skip-connection architecture that allows training very deep models efficiently. These patch embeddings were then aggregated to produce a single slide-level representation. Non-image features, such as clinical variables or genomic profiles, are usually processed with multilayer perceptrons.
When it came to integrating modalities, we found considerable variation. Studies have used early, intermediate, and late fusion approaches, but intermediate fusion is by far the most common. In early fusion, raw features are concatenated, and in late fusion, predictions from separate models are combined. Instead, intermediate fusion brings both modalities into a shared latent space, enabling joint learning. The most frequently used techniques were contrastive learning and cross-attention-based mechanisms. Only one study used early fusion, three applied late fusion, and all remaining studies used intermediate fusion.
Downstream prediction tasks focus on three main areas: survival prediction, biomarker prediction, and pathological staging. Across these applications, the results consistently showed that multimodal models outperformed single-modality approaches. WSIs provide rich morphological information, whereas clinical and genomic data contribute to patient-specific and molecular contexts. Combining these complementary sources nearly always improved the predictive performance compared to using any one modality alone.
Looking ahead
We hope our work informs both researchers and clinicians working on CRC by highlighting the research gaps, latest advances and future research directions. As part of our current research, we are experimenting with a novel contrastive-learning-based architecture on the MCO dataset and hypothesise that multimodal models can outperform single-modality approaches even with limited data. A key priority is the development of multicohort studies to improve robustness and generalisability, given the field’s current dependence on single-centre datasets. There is also major potential for adopting and adapting foundation models capable of learning richer cross-modal representations.
Looking further ahead, we see major potential in adopting and adapting foundation models that can learn richer cross-modal representations, as well as exploring agentic AI systems that can autonomously coordinate multimodal workflows, optimise models iteratively, and support scalable biomarker discovery. Finally, enhancing the explainability of multimodal systems—understanding how image and non-image features jointly contribute to predictions—remains essential for building trust and enabling real clinical use.
Follow the Topic
-
npj Digital Medicine

An online open-access journal dedicated to publishing research in all aspects of digital medicine, including the clinical application and implementation of digital and mobile technologies, virtual healthcare, and novel applications of artificial intelligence and informatics.
Related Collections
With Collections, you can get published faster and increase your visibility.
Evaluating the Real-World Clinical Performance of AI
Publishing Model: Open Access
Deadline: Jun 03, 2026
Impact of Agentic AI on Care Delivery
Publishing Model: Open Access
Deadline: Jul 12, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in