How to map the human brain (fast)?
Published in Neuroscience

Every human brain is unique, just like fingerprints or the retinas of the eyes. Aside from this structural variability, the functional organization of the cerebral cortex also varies between individuals and across one’s lifespan. In general, primary functions, such as motor control or early visual perception, have more stable relationships between the location and function of brain activity, so called structure-function relationships. In contrast, higher cognitive functions such as language require numerous sub-regions of the brain to interact and thus show a more flexible organization.
Over the last decades, new methods of neurophysiology, computational neuroscience, and advanced neuroimaging techniques have led to substantial progress in understanding the human brain. However, these studies mostly focused on average- or standard-brains by aggregating data from large group studies. Only recently have efforts been made to tap into individual cortical organization to better understand the fundamental mechanisms of brain functions in healthy and damaged brains, which is an essential prerequisite for precision medicine.
Title Figure: 3D-scan of an 1850's phrenology bust from @AnatomicalMuseum (license). TMS coil model: MagVenture MCF-B65. Background: unsplash.com. Grey matter brain surface with induced electric field strength (|E|) computed with SimNIBS.
1. Identifying causal structure-function relations with TMS
2. Cortical localization based on TMS-induced electric fields
3. Comparison with other methods
4. An interdisciplinary team effort
1. Identifying causal structure-function relations with TMS
Notably, most neuroscientific approaches merely provide correlational insights into structure-function relationships in the human brain. For example, functional magnetic resonance imaging (fMRI) can identify metabolic changes in cerebral blood flow to associate these changes with local alterations in neuronal activity. However, fMRI is not suitable to draw conclusions about the causal relationship between neuronal activity and a measured behavioral response.
In contrast to these correlative methods, non-invasive brain stimulation (NIBS) methods pursue a different approach: active modulation of neuronal activity and behavioral output. The potential to induce changes in brain functioning in a non-invasive and short-term manner render NIBS methods extremely valuable in human brain research. One example is transcranial magnetic stimulation (TMS). By applying a strong time-varying magnetic field, an electric field is induced in the brain that directly influences the stimulated neurons and thus actively modulates local and remote brain functioning. This approach has the decisive advantage of identifying a causal relationship between the induced electric field at a cortical target and an observable physiological or behavioral effect.
We took advantage of this feature and developed a novel TMS localization method to quickly, efficiently, and precisely map brain functions on the individual subject level. To do so, we constructed individual high-resolution 3D head models from structural magnetic resonance images and calculated the induced electric field distributions from TMS using state-of-the-art computation methods.
We demonstrated the high spatial resolution of this localization method by mapping individual finger muscles in the motor cortex (Weise et al. 2020, Numssen et al. 2021).
2. Cortical localization based on TMS-induced electric fields
Distinct cortical patches in the primary motor region of the brain contain neurons that control specific muscles via downstream connections to these muscles’ motor units. Stimulation of such a cortical patch with TMS yields a muscle contraction; stronger stimulation i.e., a stronger electric field of the relevant cortical patch, yields stronger contraction. However, a TMS-induced field is notoriously unfocal, exposing large parts of the cortex to substantial stimulation, and thus introduces ambiguity between observed responses and the identification of the originating brain site.
We mastered this lack of spatial specificity by combining multiple stimulations and their muscle responses into so-called cortical input-output curves (IO-curves). IO-curves represent the relationship between the induced electric field from many TMS coil positions and the measured motor responses of each stimulation (Fig. 1, Stage 2). By assessing the agreement (goodness-of-fit) between the measured data and these fitted IO-curves, we quantified the degree to which changes in cortical stimulation explain the observed motor response.
This allowed us to precisely pinpoint the cortical location that caused the motor response – by identifying the location that yielded the highest goodness-of-fit (Fig. 1, Stage 4).
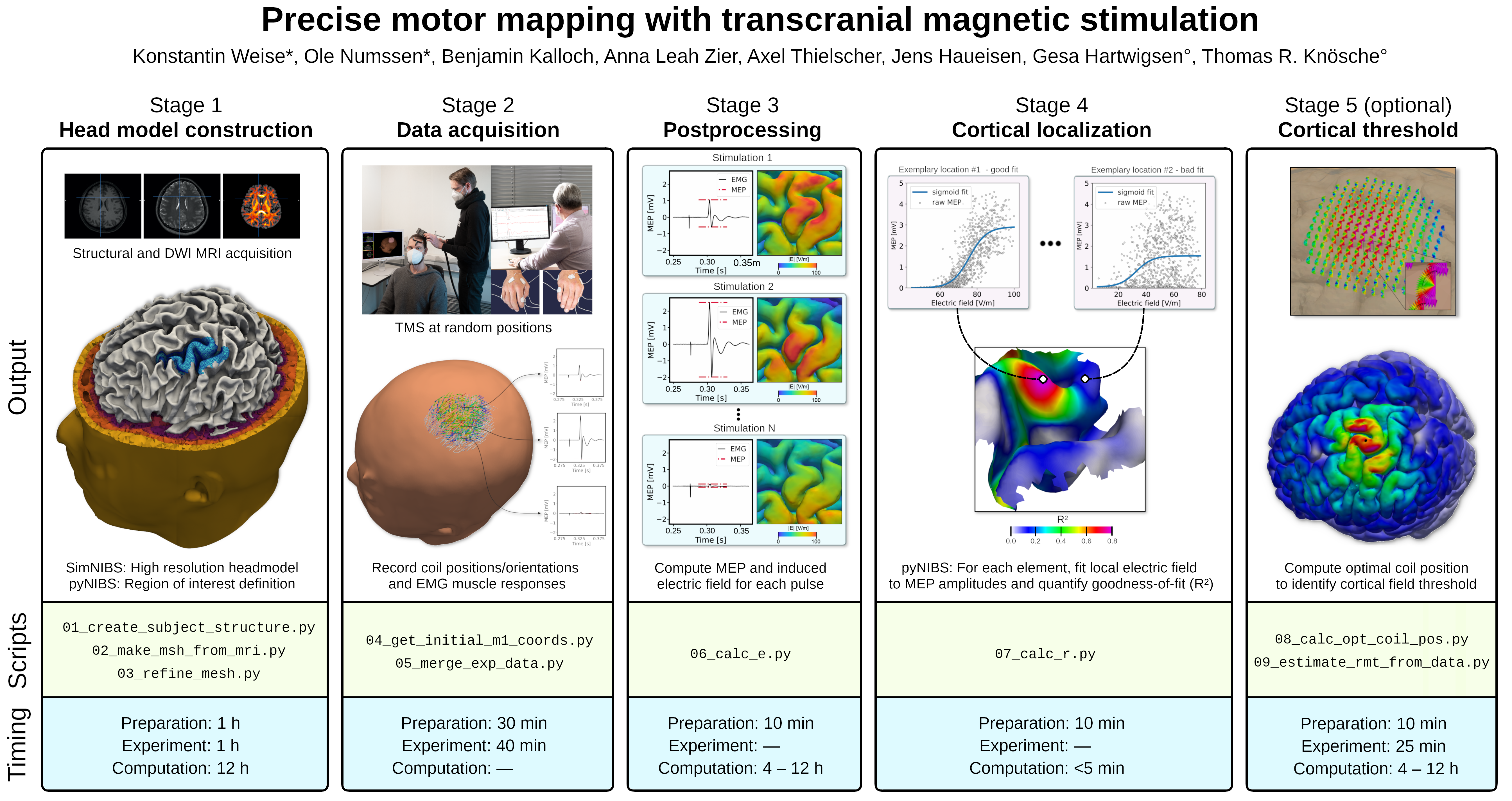
Stage 1: High-resolution head models are constructed from structural MRI images. Stage 2: Motor responses from multiple finger muscles are recorded via surface EMG for a set of quasi-random TMS coil positions/orientations. Stage 3: All TMS-induced electric fields are computed; motor evoked responses (MEPs) are extracted from the EMG data. Stage 4: The local electric field is fitted to the MEPs of each muscle to identify the cortical origins of the MEPs. Stage 5 (optional): Due to the identification of the cortical muscle representations, individual cortical field thresholds can be assessed to determine biophysiologically-plausible field-based TMS dosages. Adapted from 10.1038/s41596-022-00776-6.
In our protocol (view only), we carefully walk the reader through the process of our novel TMS mapping procedure to allow an easy application for other labs. In addition, we provide an extensive example dataset together with all relevant scripts. This Python-based analysis pipeline utilizes the NIBS simulation environment SimNIBS for the individual head-model construction (Fig. 1, Stage 1) and computation of the induced TMS electric fields (Fig. 1, Stage 3). Our previously released Python package pyNIBS is used to process the experimental data, to fit cortical IO-curves, and to identify the cortical origin of the motor response.
3. Comparison with other methods
Our proposed TMS localization method outperforms traditional mapping methods in several ways:
- Experimental duration. 100-300 stimulations each at a random position around the motor cortex suffice to yield robust and high-resolution cortical mappings, corresponding to about 10 to 25 minutes of data acquisition.
- User-independence. The TMS experiment is based on TMS pulses from quasi-random coil locations and coil orientations. The localization itself is performed post-experimentally in a fully data-driven manner and is therefore independent of the experimenter.
- Rigor. Our localization approach assesses the causal relationship between the mode-of-action of TMS, that is, the induced electric field in the cortex, and a peripheral output measurement, in this case motor evoked potentials (MEPs). This localization of functions within the cortex stands in contrast to other localization paradigms that try to localize functions based on TMS coil positions on the head and subsequent projections onto the cortical surface.
- Quality-assessment. We provide metrics to assess the quality of localizations based on the convergence of their focality, to be used when applying the localization protocol outside of the healthy population or when applying it to other functional domains (see below).
- Genericity. Our mapping protocol is – in principle – independent of the outcome measure. Any behavioral or physiological output that depends on the induced field strength can be utilized to fit cortical IO-curves, as long as a single cortical region is causally involved in this process and single-trial response data can be captured. This could, for example, also include changes in reaction times for higher cognitive tasks such as attention or language processing.
In addition to a solid identification of structure-function relationships, this method allows the assessment of individual cortical field thresholds to enable biophysiologically-plausible field-based TMS dosing (Fig. 1, Stage 5).
4. An interdisciplinary team effort
The development of this novel and advanced localization method was only possible through close interdisciplinary collaboration. Gesa Hartwigsen (Max Planck Institute for Human Cognitive and Brain Sciences, MPI CBS, Leipzig, Germany) and her Cognition and Plasticity team with Ole Numssen and Anna Leah Zier enriched our group with their expertise from the field of psychology and their experience with experimental TMS studies. Complementary, the expertise in electrical engineering of Thomas Knösche (Brain Networks Group @ MPI CBS), Konstantin Weise, and Jens Haueisen contributed significantly to the development of the algorithms to identify cause-and-effect relationships. The utilization of the well-established NIBS simulation toolbox SimNIBS through collaboration with Axel Thielscher was central to the project. Due to the focus on automatization and numerical processing, this joint effort significantly relied on Benjamin Kalloch’s background in computer science and programming skills.
However, it is not sufficient to place a group of scientists from various disciplines in a room and make them work together. They must be willing to learn to speak the same language.

With this cortical localization approach and the accompanying protocol, we strive to provide a ready-to-use framework to the NIBS community, which is based, both, on state-of-the-art methodological developments and on thorough conceptual and experimental design.
Related work
- Example dataset: 10.17605/OSF.IO/MYRQN
- Code repository: gitlab.gwdg.de/tms-localization/papers/tmsloc_proto
- Weise, K., Numssen, O., Kalloch, B., Zier, AL., Thielscher, A., Haueisen, J., Hartwigsen, G., Knösche, TR. et al. Precise motor mapping with transcranial magnetic stimulation. Nat Protoc (2022). DOI: 10.1038/s41596-022-00776-6
- Numssen, O., Zier, A. L., Thielscher, A., Hartwigsen, G., Knösche, T. R., & Weise, K. (2021). Efficient high-resolution TMS mapping of the human motor cortex by nonlinear regression. NeuroImage, 245, 118654. DOI: 10.1016/j.neuroimage.2021.118654
- Weise, K., Numssen, O., Thielscher, A., Hartwigsen, G., & Knösche, T. R. (2020). A novel approach to localize cortical TMS effects. NeuroImage, 209, 116486. DOI: 10.1016/j.neuroimage.2019.116486
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.
Your space to connect: The Psychedelics Hub
A new Communities’ space to connect, collaborate, and explore research on Psychotherapy, Clinical Psychology, and Neuroscience!
Continue reading announcement



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in