Intraoperative virtual histology
Published in Bioengineering & Biotechnology

Stimulated Raman scattering (SRS) microscopy was viewed as a major breakthrough in optics when Chris Freudiger and his colleagues described it in 20081. The following year I met Chris, a graduate student at the time, at a symposium in Stockholm where I was presenting my work on detecting tumor infiltration during brain-tumor surgery. Chris and I struck up a conversation on how SRS might be used to help surgeons detect tumor in the operating room. We discussed the guessing game that neurosurgeons must play when trying to differentiate tumor-infiltrated tissue and normal brain tissue.
At the time I was working on a dye-based approach to tumor visualization2 and struggling with finding a creative means of delivering dye specifically to tumor cells, a problem that has vexed researchers in the field for over seventy years3. To me, the idea of applying SRS to brain-tumor delineation was revolutionary because it required only the intrinsic biochemical building blocks of tissue to generate image contrast and therefore differentiate tumor infiltrated tissue from normal brain.
Shortly after meeting Chris, I was fortunate to collaborate with Minbiao Ji and other members of the Xie lab, where SRS was developed, to pursue a detailed study of SRS microscopy in the rodent brain and in rodent models of human glioma4. Our work confirmed what we had suspected—SRS could indeed reveal tumor infiltration in brain tissue that was not visible to the naked eye. Working with an outstanding team of neuropathologists, we also showed that SRS could reveal and detect key microscopic features of human brain tissue from my patients5. At the completion of these studies, there was little doubt that SRS microscopy could have a major impact on brain-tumor surgery.
However, the challenge of executing SRS microscopy in a medical environment posed a major roadblock in clinical translation. Chris led an effort to develop a novel fiber laser source capable of supporting SRS, which eliminated the safety and stability concerns associated with solid-state laser sources traditionally required for SRS microscopy6. This one-of-a-kind fiber laser is at the core of the first clinical SRS microscope described and validated in our paper.
The impact of SRS microscopy was apparent as soon as the system was delivered to our hospital. In a fraction of the 30–40 minutes required to generate and interpret conventional H&E stained slides, the SRS microscope could detect tumor infiltration and reveal key diagnostic features. Moreover, because SRS imaging requires minimal tissue preparation, the artifacts that often complicate the interpretations of conventional H&E stained slides were eliminated. To make our images easier for clinicians to interpret, we utilized a machine-learning approach to process grayscale SRS images to simulate hematoxylin and eosin images via a method that we developed, called stimulated Raman histology (SRH).
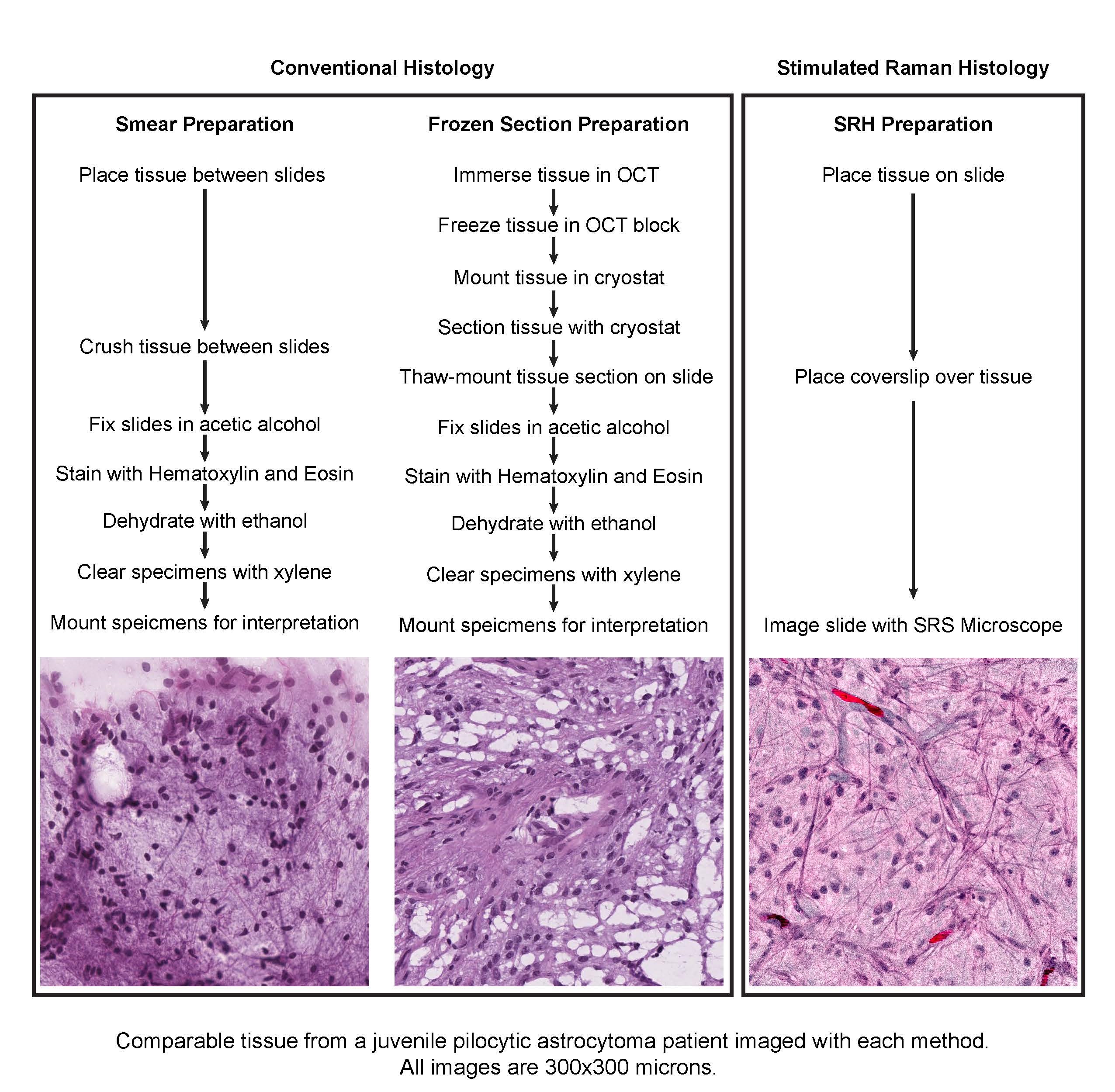
Given that SRH images are digital and fully quantifiable, we hypothesized that machine-learning techniques might be capable of assisting in intraoperative diagnosis. We generated a powerful multilayer perceptron capable of differentiating lesional and non-lesional specimens with perfect accuracy and of sorting lesional specimens into histologic subtypes with 90% accuracy. Our work provides proof of concept that machine-learning techniques can be useful for the interpretation of SRH images. Automated diagnosis via machine learning is of particular interest in the centers performing brain-tumor surgery that have limited coverage by neuropathologists.
Nine years after its discovery, SRS microscopy has now been translated from bench to bedside. SRH represents a newly validated medical-imaging method, which simplifies intraoperative tissue diagnosis and may ultimately provide safer, more effective treatment to brain-tumor patients.
Video: Intraoperative virtual histology.
Our paper: Orringer, D. A. et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng. 1, 0027 (2017).
References
1. Freudiger, C. W. et al. Label-free biomedical imaging with highsensitivity by stimulated Raman scattering microscopy. Science 322, 1857-1861, doi:322/5909/1857 [pii] 10.1126/science.1165758 (2008).
2. Orringer, D. A. et al. Small solutions for big problems: the application of nanoparticles to brain tumor diagnosis and therapy. Clin Pharmacol Ther 85, 531-534, doi:10.1038/clpt.2008.296 (2009).
3. Moore, G. Fluorescein as an agent in the differentiation of normal and malignant tissues. Science 106, 130-131 (1947).
4. Ji, M. et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med 5, 201ra119, doi:10.1126/scitranslmed.3005954 (2013).
5. Ji, M. et al. Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci Transl Med 7, 309ra163, doi:10.1126/scitranslmed.aab0195 (2015).
6. Freudiger, C. W. et al. Stimulated Raman scattering microscopy with a robust fibre laser source. Nat Photonics 8, 153-159, doi:10.1038/nphoton.2013.360 (2014).





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in